A Simplified Method for Obtaining cDNA of Low-Copy and Silent Eukaryotic Genes Using Human Renalase as an the Example
Institute of Biomedical Chemistry, 10 Pogodinskaya str, Moscow, 119121 Russia; *e-mail: valfed38@yandex.ru.
Key words: cDNA, PCR, exon, cloning, expression
DOI:10.18097/BMCRM00101
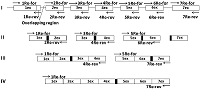
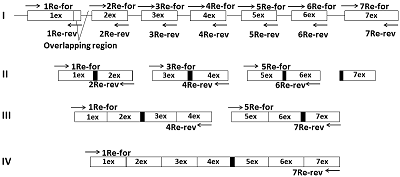
A simplified «exon» method was developed for producing cDNA of low-copy and silent eukaryotic genes. It is based on assembly of the target gene from genomic DNA by direct synthesis of its exons, followed by their PCR-based joining without further purification of the amplicons. During the synthesis of exons, direct primers were used; these included about ~ 20 nucleotides of the 3'-terminal sequence previous (from the amplified) exon and ~ 20 nucleotides of the 5'-initial sequence of the amplified exon. Reverse primers included ~ 20 nucleotides complementary to the terminal sequence of the amplified exon. Forward and reverse primers flanking the gene to be assembled included the restriction sites necessary for insertion into the expression vector. Using this approach it is possible to assemble almost any eukaryotic gene with a known nucleotide sequence of genomic DNA available in the database.
|
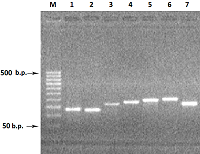
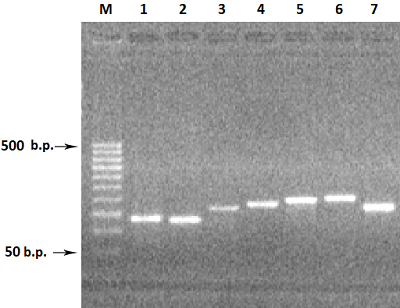
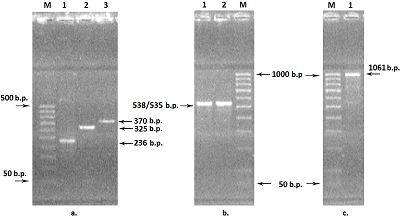
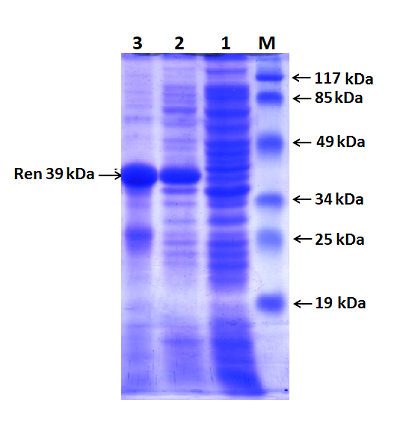
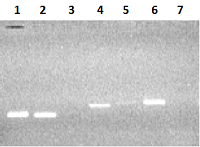
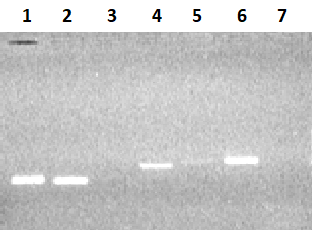
Figure 2.
Electrophoresis in 2% agarose gel of DNA (exon amplicons obtained using the genomic DNA template). The exon number corresponds to the track number in the gel.
|
|
CLOSE

|
Table 1.
Primers used in PCR amplification of 5 exons of the human renalase gene.
|
FUNDING
This study performed within the framework of the Program of Basic Scientific Research of the State Academies of Sciences for 2013–2020 was partially supported by a grant from the Russian Foundation for Basic Research No. 17–04–00484 (2017–2019).
REFERENCES
- Jayakumara, Huang, W-Y., Raetz B., Chirala ,S.S. Wakil, S.J. (1996). Cloning and expression of the multifunctional human fatty acid synthase and its subdomains in Escherichia coli. Proc. Natl. Acad. Sci. USA, 93, 14509-14514. DOI
- Dae, W.K., Won, S.E., Sang, H.J., Chang, S.Y., Young, H.K., Soo H.C., Hee S.C., So Y.K., Hyeok Y.K., Jung H.K., Oh-Shin K., Sung-Woo C., Jinseu P., Soo, Y.C. (2003). Molecular Gene Cloning, Expression, and Characterization of Bovine Brain Glutamate Dehydrogenase. BMB Reports, 36 (6), 545-551. 10.5483/BMBRep.2003.36.6.545
- Salah, E.E., Abdalla, M., Eltayb, W.A., El-Arabey, A.A., Ganash, M., Alshammari, F.D., Barreto, G., Ashraf, G.M. (2019). Molecular cloning, expression, purification, and functional characterization of SP-22 gene from Bombyx mori. J. Cell. Biochem., [Epub ahead of print] 10.1002/jcb.28826.
- Yon, J., Fried, M. (1989). Precise gene fusion by PCR. Nucleic Acids Res.; 17(12), 4895. DOI
- Ciccarelli, R.B., Gunyuzlu, P., Huang, J., Scott, C., Oakes, F.T. (1991). Construction of synthetic genes using PCR after automated DNA synthesis of their entire top and bottom strands. Nucleic Acids Res., 19(21): 6007-6013. DOI
- Haoming, G., Jose, P., Rey, G., Donella, J.W. (1991). Full length mouse glycophorin gene constructed using recombinant polymerase chain reaction. Biochem. Biophys. Res. Commun., 177 (1), 202-208. DOI
- Booth, P.M., Buchmanb, G.W., Rashtchian, A. (1994). Assembly and cloning of coding sequences for neurotrophic factors directly from genomic DNA using polymerase chain reaction and uracil DNA glycosylase. Gene, 146, 303-308. DOI
- Nisson P.C., Rashtchian A., Watkins P.C. (1991). Rapid and efficient cloning of alu-PCR products using uracil DNA glycosylase. PCR Methods Appl., 1, 120-123. DOI
- Rashtchian, A., Buchman, G., Schuster, D., Berninger M. (1992). Uracil DNA glycosylase-mediated cloning of PCR-amplified DNA: application to genomic and DNA cloning. Anal Biochem. 206, 91-97. DOI
- An, X., Lu, J., Huang, J., Zhang, B., Liu, D., Zhang, X., Zhou, Y., Tong, Y. (2007). Rapid Assembly of Multiple-Exon cDNA Directly from Genomic DNA. PloS ONE, 2(11):e1179. DOI
- Davies, W.L., Carvalho, L.S., Hunt, D.M. (2007) SPLICE: A technique for generation in vitro spliced coding sequeces from genomic DNA. BioTechniques, 43, 785-789. DOI
- Di Donato, A., de Nigris, M., Russo N., Di Biase, S., D'Alessio G. (1993). A method for synthesizing genes and cDNAs by the polymerase chain reaction. Anal Biochem. 212(1), 291-293. DOI
- Gurevich, A.I., Esipov, R.S., Kaiushin, A.L., Korosteleva, M.D. (1997) Construction of artificial genes by PCR methods using the synthetic template. Bioorgan. Khim., 23 (6), 495-496.
- Miller, S.A., Dykes, D.D., Polesky, H.F. (1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Research, 16 (3), 1215. DOI
- Grimberg, J., Nawoschik, S., Belluscio, L., McKee, R., Turck, A., Eisenberg, A. (1989). A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res., 17, 8390. DOI
- Lee, P.Y., Costumbrado, J., Hsu, C.Y., Kim, Y.H. (2012) Agarose Gel Electrophoresis for the Separation of DNA Fragments. J. Vis. Exp., 62, e3923, 10.3791/3923. DOI
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature; 227, 680-685. DOI
- Kushner, S.R. (1978). An improved method for transformation of Escherichia coli with ColE1-derived plasmids. In: Genetic engineering (Boyer H.B. and Nicosia S., eds.), p. 17, Elsevier/North-Holland, Amsterdam. DOI
- Cohen, S.N., Chang, A.C.Y., Hsu, L. (1972). Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Esherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA., 69, 2110-2114. DOI
- Mullis, K.B., Faloona, F.A. (1987) Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol., 155, 335-50. DOI
- Fedchenko, V.I., Kaloshin, A.A., Mezhevikina, L.M., Buneeva, O.A., Alexei, Medvedev A.E. (2013) Construction of the Coding Sequence of the Transcription Variant 2 of the Human Renalase Gene and Its Expression in the Prokaryotic System. Int. J. Mol. Sci., 14, 12764-12779. DOI
- Fedchenko, V., Kopylov, A., Kozlova, N., Buneeva, O., Kaloshin, A., Zgoda, V., Medvedev, A. (2016). Renalase Secreted by Human Kidney НЕК293Т Cells Lacks its N-Terminal Peptide: Implications for Putative Mechanisms of Renalase Action. Kidney Blood Press Res, 41, 583-593. DOI