|
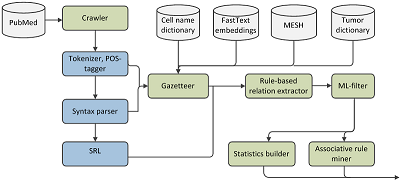
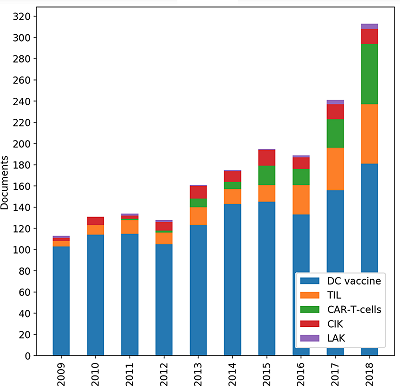
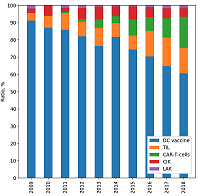
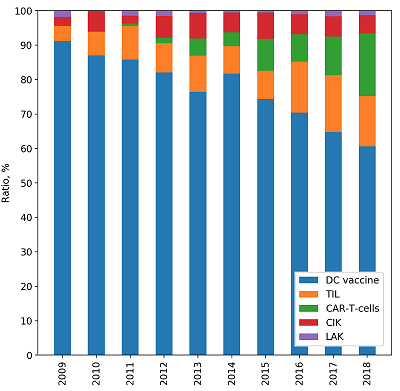
CONTENTS COMPLIANCE WITH ETHICAL STANDARDS Figure 1 The framework for meta-analysis of biomedical texts. Figure 2 Dynamic distribution of the documents devoted to particular cell-based immunotherapies. Figure 3The ratios of the documents devoted to particular cell-based immunotherapies. Table 1Categories, queries and counts of the extracted documents. Table 3Associative patterns for cell-based immunotherapy of cancer. Table 4Evaluation results for the proposed framework. |
Towards Automated Meta-Analysis of Biomedical Texts on the Field of Cell-Based Immunotherapy
1 Federal Research Centre «Computer Science and Control» RAS, Key words: cancer; cell-based immunotherapy; text mining; automated meta-analysis Abbreviations: ADCC – antibody dependent cell-mediated cytotoxicity; APC – antigen presenting cells; AP B-cells – antigen-presenting B-cells; CAR – chimeric antigen receptor; CIK – cytokine-induced killers; DC – dendritic cells; LAK – lymphokine activated killers; MHC – main histocompatibility complex; NER – named entities; NK-cells – natural killer cells; NLP – natural language processing; TCR – T-cell receptor; TIL – tumor-infiltrating lymphocytes DOI: 10.18097/BMCRM00109 INTRODUCTION Cell-based immunotherapy is a rapidly growing area of biomedical science aimed at cell transplantation to achieve a therapeutic effect. Cells exhibiting immunomodulating properties can be used to suppress pathological immune responses to autoantigens, as well as to stimulate immunity to fight against tumor or chronic infections. To treat cancer by administration of immune cells two main approaches are actively investigated in research laboratories and clinics. The first approach employs active immunotherapy with a cell vaccine, which can present tumor antigens to lymphocytes and induce an immune response. It is based on the use of antigen presenting cells (APC) or inactivated tumor cells. The most effective APC are dendritic cells (DC) that can present tumor antigens in the context of their own main histocompatibility complex (MHC) molecules to lymphocytes [1]. The usual procedure of the immunotherapy incudes isolation of monocytes from patients’ blood, their ex vivo differentiation to DC, loading DC with a tumor antigen followed by DC growth and maturation in vitro, administration of antigen loaded DC to the patient. Alternatively, DC can be fused in vitro with tumor cells to produce dendritomas. The second approach is based on a passive (adoptive) immunization [2]. It includes injections of various immune effector cells, usually lymphocytes, capable of lysing tumor cells and isolated from blood or tumor (tumor-infiltrating lymphocytes (TIL)). The lymphocytes must be propagated and «trained » in a culture to achieve the therapeutic effect. To direct lymphocytes against a tumor, various cytokines (lymphokine activated killer (LAK), cytokine-induced killer (CIK)) or a genetic modification of antigen-recognizing receptors (redirected TCR T-cells (reTCR T-cells), chimeric antigen receptor (CAR) T-cells) can be used. Usually any procedure for conducting cell-based immunotherapy includes isolation of immune cells from a patient, their modification and propagation in a cell culture, and subsequent administration back to the patient. However, it is rather difficult to track development and evaluate effectiveness of cell-based immunotherapy methods by analyzing scientific literature. There are a large number of cell populations with immunomodulatory properties and they can be cultured under various conditions to improve their therapeutic characteristics. In addition, the characteristics of the patient and patient’s disease can significantly affect outcome of the treatment. The problem is complicated by the fact that there are thousands of texts related to this topic. In this regard, development of a tool for automated meta-analysis of biomedical texts in the field of cell-based immunotherapy is a challenge facing cell biology and computer science. The automated meta-analysis consists in retrieving of required research papers, accurate extraction of particular named entities (NER), revealing their roles and relations between them. Complex linguistic features are usually required to tackle with these problems. Krallinger with colleagues highlighted the natural language processing (NLP) methods to retrieve some of these features [3]. There are specific NLP components for biomedical texts such as POS-taggers [4] and syntax parsers [5], which were trained on specific domain models to return syntactic dependency relations between tokens in a clause. The approaches for bio-NER include dictionary-lookup [6], rule-based [7], and machine-learning-based sequence labeling [8]. Nevertheless, the quality of all these techniques depends on the entity type, its linguistic features and availability of dictionaries. It is worth to note the following supervised machine learning models applied to detect biomedical entities: SVM [9], conditional random field (CRF) [10], and recurrent neural networks [11]. Besides good quality of extraction these models require large corpora to train reliably; however, this represents a serious problem due to limitation in the the existing data for information extraction. In the case of cell-based therapy, we should extract mentions of specific cell types, their roles in the text and particular types of relationships between them. At the same time, we do not have any labeled corpora applicable to training end-to-end machine learning approaches. Considering this situation, a hybrid approach has been proposed. Our solution consists in applying gazetteers and rule-based analyzer together to detect candidates for the entities and then to filter them with a pre-trained machine learning model. We also consider all available external linguistic resources, such as MESH (https://meshb.nlm.nih.gov/search), pre-trained FastText models [12], cell and tumor dictionaries to tackle with the small corpora problem. After that syntax and semantic features are used to reveal relations between the entities. A similar approach was employed by Shelmanov and colleagues [13]. In this paper, we have proposed a framework, which provides solutions for NLP tasks that arise in meta-analysis: from web-crawling to named entities recognition and estimation of statistic scores. This approach has been applied to identify associations between types of tumors and the most commonly used methods of their cell-based immunotherapy. METHODS As a basic method for the analysis, we propose a framework that contains several steps (Fig. 1). 1. Crawling abstracts from Pubmed. We applied Scrapy- based web-crawler with a manually created rule set to download the search output from PubMed [14]. 2. Rich linguistic features extraction. In this step the Isanlp framework is applied to obtain morphology, syntax parsing, and semantic role labeling features [15]. 3. Combining tumor and cell dictionaries (provided by biologists) and morphology-based rules to extract entity candidates from the abstracts. We use syntax relations and build all their possible combinations. Regular expression- based features were also considered. This is an adaptation of the linguistically motivated approach for mapping terms from medical texts to concepts in UMLS Metathesaurus, which was proposed in https://www.nlm.nih.gov/research/umls/knowledge_ sources/metathesaurus/index.html and called MetaMap [7]. We also applied the Fasttext model, pre-trained on biomedical texts, to catch synonyms. 4. Syntax and semantic features analysis. We use syntax relations and semantic roles to reveal links between the entities and their roles (target cell, active cell) in the sentence. 5. Applying a pre-trained sequence-labeling machine learning model to filter uninformative entity candidates. This step has not been implemented yet, now we use dictionary-based filter instead. 6. Coocurence statistics calculating and associative rule mining for the extracted entities [16] to obtain stable combinations of tumors, therapy and cell types. We used the Eclat algorithm [17] because of its scalability. We also created a lexis only approach for the steps 3 – 4, which do not consider any syntax or semantic features. This approach is used as a baseline in the experiments on text mining.
RESULTS AND DISCUSSION In the frst instance, we extracted 96160 non-labeled abstracts related to cell-based cancer immunotherapy from PubMed (Table 1). The total corpus of the texts did not include annotations of reviews and was divided into three groups for separate analysis. The frst group contained experimental research texts on a human material including established cell lines and patients’ tissue samples. The second group integrated similar animal studies and preclinical trials. The third group corresponded to clinical trials conducted according to the relevant regulations. Separate analysis was useful in assessing the maturity of immunotherapeutic approaches.
Based on the thesaurus of cell-based immunotherapy methods compiled by specialists and applying proposed framework we extracted the data on various immunotherapy methods from the primary text corpus (Table 2).
The data demonstrate that DC vaccines are the most common method of immunotherapy for both basic researchers and clinical trials. Despite rather low effciency [1], high impact of DC vaccines is apparently due to the simplicity of their preparation and the absence of side effects in patients. Obviously, the efforts of researchers in this direction is mainly aimed at increasing the effectiveness of DC vaccination. Dynamic analysis shows that although there is growth in the number of studies on DC vaccines (Fig. 2), their percentage among other methods is gradually decreasing (Fig. 3). More and more methods of passive immunotherapy, including genetically modifed effector cells, are entering the stage of clinical trials. Interestingly, cell-based immunotherapy approaches with a long history, such as TIL or CIK, are still involved in clinical trials. Perhaps this is because of the recent opportunity to signifcantly increase the effectiveness of passive cell-based immunotherapy by applying lymphodepleting chemotherapy before the cell administration [18]. In addition, the development of methods for propagating in vitro antigen-specifc lymphocyte clones has improved the clinical outcome of traditional approaches [19].
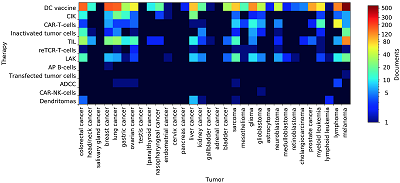
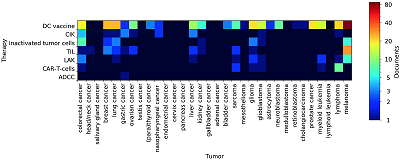
An important part of the study is the identifcation of associative relationships between cell-based immunotherapy methods and the tumors for which the treatment is intended. We applied a dictionary of tumor names to both the whole corpus of documents and the clinical trial documents subset. DC vaccines is mentioned by researchers in the context of almost all of tumor types (Fig. 4). In clinical studies, DC loaded with tumor antigens are most commonly used to treat melanoma, prostate cancer, brain tumors, colorectal cancer, lung cancer, breast cancer, and lymphomas (Fig. 5). Quite interesting data are related to the immunotherapy with CAR-T-cells. A feature of such effector cells is that their chimeric antigen-recognizing receptor interacts with a tumor antigen that is not associated with MHC on the tumor surface. Since the presence of such kind of antigens is mostly a characteristic feauture of hematopoietic neoplasms, B-cell lymphomas expressing CD19 antigen have always been a common target for CAR-T-cells [20]. Our analysis has shown that lymphomas are the main target for CAR-T-cells based therapy (Fig. 5). However, there are many mentions of this effector in the context of solid tumors (Fig. 4) and even clinical trials, mainly aimed at treating sarcomas and glioblastomas (Fig. 5). It indicates the development of CAR-Tcells based therapy towards the treatment of solid tumors. Table 3 presents the most confdent associations between the methods of immunotherapy and tumor types mined from analyzed abstracts.
To evaluate the effectiveness of our approach to automated meta-analysis we have tested the proposed framework against baseline approach on a small corpus, which contains 16 manually labeled abstracts from PubMed Central (Table 4). Through the experiment, we estimated precision (P), recall (R) and F1 scores as the most important characteristics [21]. The results show that the proposed framework significantly over performs the baseline approach, especially in terms of precision. It means that syntax and semantic features are crucial for this task. However, the lower recall scores display that an additional extension of the dictionaries is still needed.
The main feature of our solution is considering all available external linguistic resources, such as cell and tumor dictionaries, pre-trained FastText embeddings and ontologies, together with the results of full linguistic analysis. This allows significantly reducing demand on the size of training corpora. In contrast, the recent deep neural-network-based language models, like a BERT, are usually pre-trained to extract complex features in an unsupervised manner and then fine-tuned in large labeled corpora to solve a particular problem; therefore external resources are hardly considered. Although these models demonstrate astonishing results in the biomedical domain [22], the demand on the size of labeled data remains high for these models. For example, it is demonstrated that about 2 000 texts is required to train such model even with transfer learning [23]. Unfortunately, the existing large corpora often do not meet the aim of a particular meta-analysis; they usually focus on general tasks. Hence, this approach is not applicable in our case. Another feature of the proposed approach is that it provides an end-to-end solution for the meta-analysis problem. In spite of plenty of studies, which are related to particular information extraction tasks, there is a lack of full information extraction frameworks for biomedical purposes. It is worth noticing that the only similar framework has been proposed by Hakala with colleagues [24]. Thus, in this article, we have shown that the proposed framework can be used as a tool for automated meta-analysis of biomedical texts in the field of cell-based immunotherapy. Moreover, if the modules for assessment of clinical outcomes [25, 26] were added to this framework, it would be useful for the generalizing meta-analysis of clinical trials effectiveness of cell-based immunotherapy. COMPLIANCE WITH ETHICAL STANDARDS We did not use biological samples from humans or other animals in this work. The analysis of clinical trial did not include patient personal data. FUNDING This study was supported by the Russian Foundation for Basic Research, grants no. 16-29-07246 and 16-29-07210. REFERENCES
|