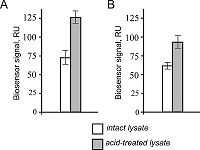
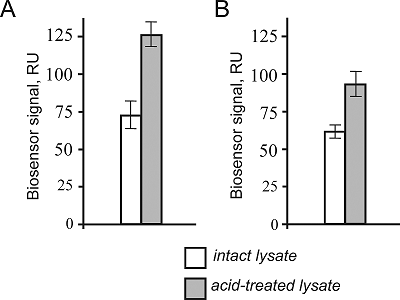
Quality Control of FITC-Labeled Proteins for Interactomics Investigations Using SDS Capillary Gel Electrophoresis and SPR Biosensorg
Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 119121 Russia; *e-mail: evgeyablokov1988@mail.ru
Keywords:surface plasmon resonance (SPR); capillary gel electrophoresis; interactomics; cytochrome c; dye labeling; FITC
DOI:10.18097/BMCRM00112
29 conjugates of methylene blue and four chemical structures, including derivatives of carbazole, tetrahydrocarbazole, substituted indoles and γ-carboline, combined with a 1-oxopropylene spacer have been studied as channel blockers of the NMDA receptor (binding site of MK-801) by using four QSAR methods (multiple linear regression, random forest, support vector machine, Gaussian process) and molecular docking. QSAR models have satisfactory characteristics. The analysis of regression models at the statistical level revealed an important role of the hydrogen bond in the complex formation. This was also confirmed by the study of modeled by docking complexes. It was found that the increase in the inhibitory activity of the part of compounds could be attributed to appearance of additional H bonds between the ligands and the receptor.
FUNDING
Capillary electrophoresis study was supported by the Russian Foundation for Basic Research (RFBR) (project No 18-04-00071).
REFERENCES
- Florinskaya, A.V., Ershov, P.V., Mezentsev, Y.V., Kaluzhskiy, L.A., Yablokov, E.O., Buneeva, O.A., Zgoda, V.G., Medvedev, A.E., Ivanov, A.S. (2018) The analysis of participation of individual proteins in the protein interactome formation. Biomeditsinskaya Khimiya, 64(2), 169–174. DOI
- Ershov, P.V., Mezentsev, Y.V., Yablokov, E.O., Kaluzhskiy, L.A., Vakhrushev, I.V., Gnedenko, O.V., Florinskaya, A.V., Gilep, A.A., Usanov, S.A., Yarygin, K.N., Ivanov, A. S. (2019) A Method of Lysate Preparation to Improve the Isolation Efficiency of Protein Partners for Target Proteins Encoded by the Genes of Human Chromosome 18. Biomed. Chem. Res. Methods, 2(1), e00090. DOI
- Ershov, P.V., Mezentsev Y.V., Kopylov, A.T., Yablokov, E.O., Svirid, A.V., Lushchyk, A.Ya., Kaluzhskiy, L.A., Gilep, A.A., Usanov, S.A., Medvedev, A.E. and Ivanov, A.S. (2019) Affinity Isolation and Mass Spectrometry Identification of Prostacyclin Synthase (PTGIS) Subinteractome. Biology (Basel), 8(2), 49. DOI
- Ng, S., Smith, M.B., Smith, H.T., Millett, F. (1977) Effect of modification of individualcytochrome c lysines on the reaction with cytochrome b5. Biochemistry, 16(23), 4975-4978. DOI
- Arrell, M.S., Kálmán, F. (2016) Estimation of protein concentration at high sensitivityusing SDS-capillary gel electrophoresis-laser induced fluorescence detection with 3-(2-furoyl)quinoline-2-carboxaldehyde protein labeling. Electrophoresis, 37(22), 2913-2921. DOI
- Ershov, P.V., Mezentsev, Y.V., Yablokov, E.O., Kaluzhskiy, L.A., Florinskaya, A.V., Gnedenko, O.V., Zgoda, V.G., Vakhrushev, I.V., Raeva, O.S., Yarygin, K.N., Gilep, A. A., Usanov, S. A., Medvedev, A. E., Ivanov, A. S. (2018) Direct Molecular Fishing of Protein Partners for Proteins Encoded by Genes of Human Chromosome 18 in HepG2 Cell Lysate. Russ. J. Bioorg. Chem., 44, 759–776. DOI