Optimization of the Protocol for Isolation of Exosome and Secretome of Caco-2 Colorectal Adenocarcinoma Cells from Conditioned Media for Subsequent Mass Spectrometric Analysis
Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 119121 Russia; *e-mail: n.shushkova@yandex.ru
Keywords:exosomes; secretion; extracellular vesicles; mass spectrometry; proteomics
DOI:10.18097/BMCRM00118
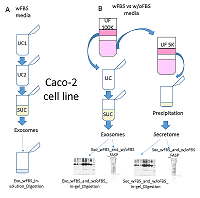
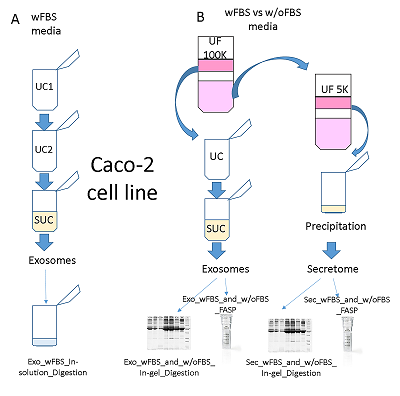
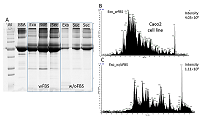
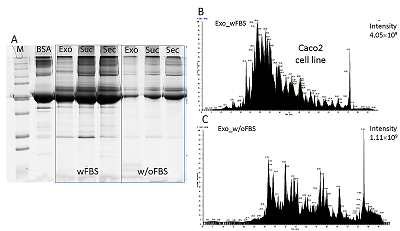
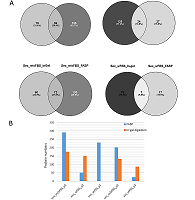
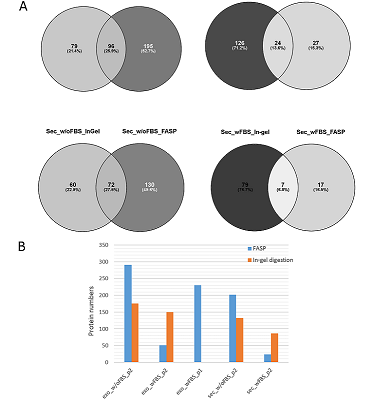
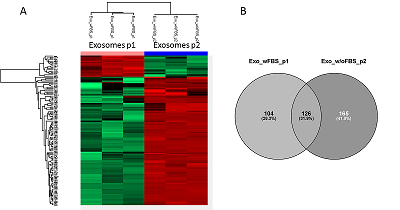
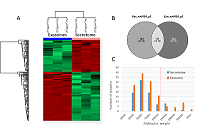
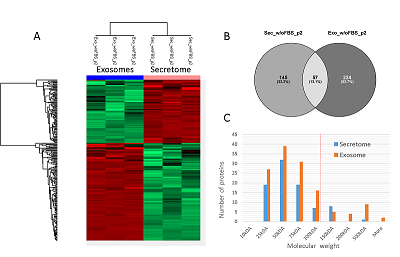
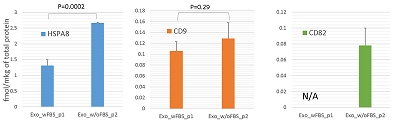
The conditioned media in which the tumor cells are cultured represents a model object for studying the secretome and proteomic composition of exosomes. This pool of proteins is of particular interest in terms of the search for potential markers of malignant diseases. The efficiency of the isolation of exosomes and secretome from the cell culture media largely determines the success of their analysis by the mass spectrometric method. In this paper, we have applied various approaches to isolate secretome and exosomes originated from Caco-2 colorectal adenocarcinoma cells. The combination of ultrafiltration and centrifugation provided a deep proteomic analysis of the secretome and exosomes obtained from the one initial volume of conditioned medium without the addition of fetal bovine serum. Applying tandem mass spectrometric analysis we have identified 436 secreted proteins. In exosomes, the characteristic proteins ALIX, CD63, syntenin, lactadherin (MFGE8) were identified. Using targeted mass spectrometry, the exosome markers CD82, CD9 and HSPA8 were determined at the levels of 0.08 ± 0.03 fmol/μg, 0.13 ± 0.03 fmol/μg and 2.6 ± 0.02 fmol/μg of total protein, respectively. Among the secreted proteins, there were many markers associated with tumor progression and metastasis, such as DAG1, PODXL, LRRN4, TGFBI, IL6ST, DSC1, DSG1, and NPC2.
|
CLOSE

|
Table 1.
The number of proteins identified in exosome and secretome samples of Caco-2 cell line.
|
FUNDING
The work was performed in the framework of the Program for Basic Research of State Academies of Sciences for 2013-2020.
SUPPLEMENTARY
Supplementary materials are available at http://dx.doi.org/10.18097/BMCRM00118
REFERENCES
- Bettger W.J., McKeehan W.L. (1986) Mechanisms of cellular nutrition. Physiol. Rev., 66(1), 1-35. DOI
- Severino V., Farina A., Chambery A. (2013) Analysis of secreted proteins. Methods Mol. Biol., 1002, 37-60. DOI
- Bosse K., Haneder S., Arlt C., Ihling C.H., Seufferlein T., Sinz A. (2016) Mass spectrometry-based secretome analysis of non-small cell lung cancer cell lines. Proteomics, 16(21), 2801-2814. DOI
- Lobb R.J., Becker M., Wen S.W., Wong C.S., Wiegmans A.P., Leimgruber A., Möller A. (2015) Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles, 4, 27031. DOI
- Wessel D., Flügge U.I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem, 138(1), 141-143.
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem, 68(5), 850-858.
- Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods, 6(5), 359-362. DOI
- Shushkova N.A., Vavilov N.E., Novikova S.E., Farafonova T.E., Tikhonova O.V., Liao P.C., Zgoda V.G. (2018) Quantitative proteomics of human blood exosomes. Biomed. Khim. 64(6), 496-504. DOI
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics, 13(9), 2513-2526. DOI
- Kopylov A.T., Ponomarenko E.A., Ilgisonis E.V., Pyatnitskiy M.A., Lisitsa A.V., Poverennaya E.V., Kiseleva O.I., Farafonova T.E., Tikhonova O.V., Zavialova M.G., Novikova S.E., Moshkovskii S.A., Radko S.P., Morukov B.V., Grigoriev A.I., Paik Y.K., Salekdeh G.H., Urbani A., Zgoda V.G., Archakov A.I. (2018) Targeted Quantitative SRM SIS Screening of Chromosomes 18, 13, Y, and the Mitochondrial Chromosome Encoded Proteome. J. Proteome Res. American Chemical Society, 18(1), 120-129, DOI
- Jeppesen D.K., Nawrocki A., Jensen S.G., Thorsen K., Whitehead B., Howard K.A., Dyrskjot L., Orntoft T.F., Larsen M.R., Ostenfeld M.S. (2014) Quantitative proteomics of fractionated membrane and lumen exosome proteins from isogenic metastatic and nonmetastatic bladder cancer cells reveal differential expression of EMT factors. Proteomics, 14(6), 699–712, DOI
- Shushkova N.A., Novikova S.E., Zgoda V.G. (2019) Exosomes of malignant tumors: prospects of omiсs diagnostics. Biomeditsinskaya khimiya, 65(6), 457-467.
- Braun F., Bertin-Ciftci J., Gallouet A.S., Millour J., Juin P., (2011) Serum-nutrient starvation induces cell death mediated by bax and puma that is counteracted by p21 and unmasked by Bcl-x L inhibition. PLoS One, 6(8), DOI
- García-Lorenzo A., Rodríguez-Pineiro A.M., Rodríguez-Berrocal F.J., Cadena M.P., Martínez-Zorzano V.S. (2012) Changes on the Caco-2 secretome through differentiation analyzed by 2-D differential in-gel electrophoresis (DIGE). Int. J. Mol. Sci., 13(11), 14401–14420, DOI