Potential Inhibitors of Protease 3CLpro Virus COVID-19: Drug Reposition
1Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 119121 Russia; *e-mail: veselov@ibmh.msk.su
2Institute of Physiologically Active Compounds, Chernogolovka 142432, Russia
Keywords:COVID-19; protease, inhibitor; drug reposition; docking; molecular dynamics
DOI:10.18097/BMCRM00124
Pneumonia caused by the COVID-19 virus has led to quick search of drugs that would able to block the spread of this virus. A standard way of drug development is a long process. One approach that can significantly accelerate drug development is drug reposition. In this study a virtual screening of the database of approved drugs has been used for search inhibitors against 3СLpro COVID-19, the main protease of COVID-19. Molecular docking, simulation of molecular dynamics and binding energy estimation by MM-GBSA method allowed to select several compounds for further experimental testing. The most promising drugs are the HIV protease inhibitor Indinavir, the inhibitor of protease hepatitis C Telaprevir, the antiulcer drug Dalargin, and the ErB receptor tyrosine kinase inhibitor Neratinib

|
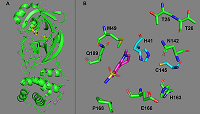
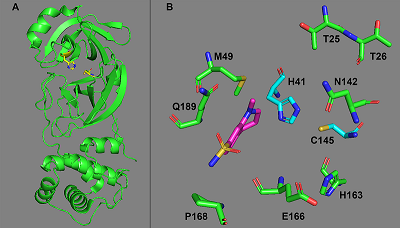
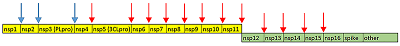
Figure 1.
Cleavage sites of COVID-19 proteases PLpro (blue arrows) and 3CLpro (red arrows). pp1a is in yellow, pp1ab – green.
|
|
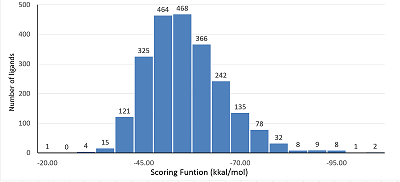
Figure 3.
A distribution of quantity of ligands depend of SFD values during docking in protease 3CLpro COVID-19. The best docking solution was used for plotting.
|
|
CLOSE

|
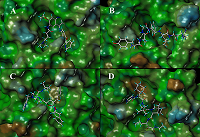
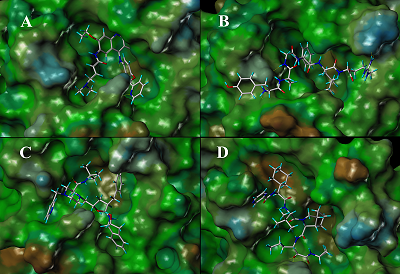
Table 1.
Drug selected as potential inhibitors of 3СLpro COVID-19 and values of their binding.
|
|
CLOSE

|
Table 2.
The main pharmacological activity of selected drugs for 3СLpro COVID-19.
|
FUNDING
The work was performed in the framework of the Program for Basic Research of State Academies of Sciences for 2013-2020.
REFERENCES
- Cui, J., Li, F., Shi, Z.L. (2019) Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol., 17(3), 181-192. DOI
- de Wit, E., van Doremalen, N., Falzarano, D., Munster, V.J. (2016) SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol., 14(8), 523-534. DOI
- Zhou, P., Yang, X.L., Wang, X.G., Hu, B., Zhang, L., Zhang, W., Si, H.R., Zhu, Y., Li, B., Huang, C.L., Chen, H.D., Chen, J., Luo, Y., Guo, H., Jiang, R.D., Liu, M.Q., Chen, Y., Shen, X.R., Wang, X., Zheng, X.S., Zhao, K., Chen, Q.J., Deng, F., Liu, L.L., Yan, B., Zhan, F.X., Wang, Y.Y., Xiao, G.F., Shi, Z.L. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270-273. DOI
- Wu, F., Zhao, S., Yu, B., Chen, Y.M., Wang, W., Song, Z.G., Hu, Y., Tao, Z.W., Tian, J.H., Pei, Y,Y., Yuan, M.L., Zhang, Y.L., Dai, F.H., Liu, Y., Wang, Q.M., Zheng, J.J., Xu, L., Holmes, E.C., Zhang, Y.Z. (2020) A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265-269. DOI
- Hegyi, A., Ziebuhr, J. (2002) Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol.. .83(Pt 3), 595-599. DOI
- Pillaiyar, T., Manickam, M., Namasivayam, V., Hayashi, Y., Jung, S.H. (2016) An Overview of Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) 3CL Protease Inhibitors: Peptidomimetics and Small Molecule Chemotherapy. J. Med. Chem., 59(14), 6595-6628. DOI
- Gautret, P., Lagier, J.C., Parola, P., Hoang, V.T., Meddeb, L., Mailhe, M., Doudier, B., Courjon, J., Giordanengo, V., Vieira, V.E., Dupont, H.T., Honoré, S., Colson, P., Chabrière, E., La Scola, B., Rolain, J.M., Brouqui, P., Raoult, D. (2020) Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents, 105949. DOI
- Novac, N. (2013) Challenges and opportunities of drug repositioning. Trends Pharmacol. Sci., 34(5), 267-272. DOI
- Jin, G., Wong, S.T. (2014) Toward better drug repositioning: prioritizing and integrating existing methods into efficient pipelines. Drug Discov. Today, 19(5), 637-644. DOI
- Pushpakom, S., Iorio, F., Eyers, P.A., Escott, K.J., Hopper, S., Wells, A., Doig, A., Guilliams, T., Latimer, J., McNamee, C., Norris, A., Sanseau, P., Cavalla, D., Pirmohamed, M. (2019) Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov., 18(1), 41-58. DOI
- Ashburn, T.T., Thor, K.B. (2004) Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov., 3(8), 673-683. DOI
- Gupta, S.C., Sung, B., Prasad, S., Webb, L.J., Aggarwal, B.B. (2013) Cancer drug discovery by repurposing: teaching new tricks to old dogs. Trends Pharmacol. Sci., 34(9), 508-517. DOI
- Tobinick, E.L. (2009) The value of drug repositioning in the current pharmaceutical market. Drug News Perspect., 22(2), 119-125. DOI
- Baek, M.C., Jung, B., Kang, H., Lee, H.S., Bae, J.S. (2015) Novel insight into drug repositioning: Methylthiouracil as a case in point. Pharmacol. Res., 99, 185-193. DOI
- Khan, R.J., Jha, R.K., Amera, G., Jain, M., Singh, E., Pathak, A., Singh, R.P., Muthukumaran, J., Singh, A.K. (2020) Targeting SARS-CoV-2: A Systematic Drug Repurposing Approach to Identify Promising Inhibitors Against 3C-like Proteinase and 2'-O-RiboseMethyltransferase. J. Biomol. Struct. Dyn. DOI
- Kandeel, M., Al-Nazawi, M. (2020) Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. DOI
- Ortega, J.T., Serrano, M.L., Pujol, F.H., Rangel, H.R. (2020) Unrevealing sequence and structural features of novel coronavirus using in silico approaches: The main protease as molecular target. EXCLI J., 19, 400-409. DOI
- Shah, B., Modi, P., Sagar, S.R. (2020) In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sci., 252, 117652. DOI
- Balius, T.E., Mukherjee, S., Rizzo, R.C. (2011) Implementation and evaluation of a docking-rescoring method using molecular footprint comparisons. J. Comput. Chem., 32(10), 2273-2289. DOI
- Humphrey, W., Dalke, A., Schulten, K. (1996) VMD: visual molecular dynamics. J. Mol. Graph., 14(1),:33-38. DOI
- Ghosh, A.K., Osswald, H.L., Prato, G. (2016) Recent Progress in the Development of HIV-1 Protease Inhibitors for the Treatment of HIV/AIDS. J. Med. Chem., 59(11), 5172-5208. DOI
- McCauley, J.A., Rudd, M.T. (2016) Hepatitis C virus NS3/4a protease inhibitors. Curr. Opin. Pharmacol., 30, 84-92. DOI
- Retrieved April 7, 2020, from: http://fmbaros.ru/press-tsentr/novosti/detail/?ELEMENT_ID=38187
- Singh, J., Petter, R.C., Baillie, T.A., Whitty, A. (2011) The resurgence of covalent drugs. Nat. Rev. Drug Discov., 10(4), 307-317. DOI