Processing Oxford Nanopore Long Reads Using Amazon Web Services
1Center for Strategic Planning and Management of Medical and Biological Health Risks,
10 bldg 1 Pogodinskaya str., Moscow, 119121 Russia
2Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 119121 Russia; *e-mail: g-s2011@mail.ru
3West Siberian Interregional Scientific and Educational Center, Tyumen State University,
6 Volodarsky str., Tyumen, 625003 Russia
Keywords:postgenomic technologies, transcript; RNA, sequencing; bioinformatics; cloud computing
DOI:10.18097/BMCRM00131
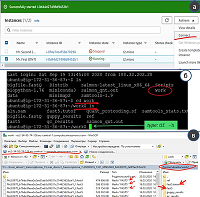
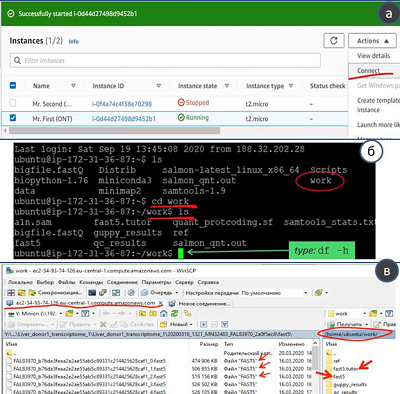
Studies of genomes and transcriptomes are performed using sequencers that read the sequence of nucleotide residues of genomic DNA, RNA, or complementary DNA (cDNA). The analysis consists of an experimental part (obtaining primary data) and bioinformatic processing of primary data. The bioinformatics part is performed with different sets of input parameters. The selection of the optimal values of the parameters, as a rule, requires significant computing power. The article describes a protocol for processing transcriptome data by virtual computers provided by the cloud platform Amazon Web Services (AWS) using the example of the recently emerging technology of long DNA and RNA sequences (Oxford Nanopore Technology). As a result, a virtual machine and instructions for its use have been developed, thus allowing a wide range of molecular biologists to independently process the results obtained using the "Oxford nanopore".
|
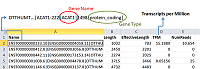
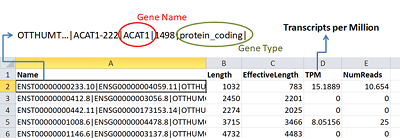
Figure 3.
The loading of data about the quantitative content of protein-coding gene transcripts in a biosample from quant.sf file to Microsoft Excel spreadsheet.
|
|
CLOSE

|
Table 1.
The configuration parameters of virtual machines to process data obtained using ONT. vCPU - virtual CPU, GPU - NVidia graphic processor unit.
|
|
CLOSE

|
Table 2.
The computing time required to process the results of human transcriptome analysis using nanopore ONT sequencing technology on Amazon Web Services.
|
|
CLOSE

|
Table 3.
The size of raw data files and files with result of processing. Estimation time of downloading the results from the AWS server.
|
FUNDING
This work was supported by the Program of Fundamental Scientific Research of State Academies of Sciences for 2013-2020.
SUPPLEMENTARY
Supplementary materials are available at http://dx.doi.org/10.18097/BMCRM00131
REFERENCES
- Van der Auwera, G. A., O’Connor, B. D. (2020) Genomic in the Cloud: Using Docker, GATK, and WDL in Terra.
- Tyanova, S., Temu, T., Cox, J. (2016) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc, 11(12), 2301–19. DOI
- Forsberg, E. M., Huan, T., Rinehart, D., Benton, H. P., Warth, B., Hilmers, B., Siuzdak, G. (2018) Data processing, multi-omic pathway mapping, and metabolite activity analysis using XCMS Online. Nat. Protoc, 13(4), 633–51. DOI
- Li, B., Dewey, C. N. (2011) RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12, 323(2011) DOI
- Langmead, B., Trapnell, C., Pop, M., Salzberg, S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol., 10, R25(2009). DOI
- Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., Lesin, V. M., Nikolenko, S. I., Pham, S., Prjibelski, A. D., Pyshkin, A. V., Sirotkin, A. V., Vyahhi, N., Tesler, G., Alekseyev, M. A., Pevzner, P. A. (2012) SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol., 19(5), 455–77. DOI
- Direct RNA Sequencing. Oxford Nanopore Technologies. Retrieved September 1, 2020, from: https://store.nanoporetech.com/media/wysiwyg/pdfs/SQK-RNA002/Direct_RNA_sequencing_SQK-RNA002_-minion.pdf
- Ilgisonis, E., Lisitsa, A., Kudryavtseva, V., Ponomarenko, E. (2018) Creation of Individual Scientific Concept-Centered Semantic Maps Based on Automated Text-Mining Analysis of PubMed. Adv Bioinformatics, 2018, 4625394. DOI
- Boža, V., Perešíni, P., Brejová, B., Vinař, T. (2020) DeepNano-blitz: a fast base caller for MinION nanopore sequencers. Bioinformatics, 36(14), 4191–4192. DOI
- Makałowski, W., Shabardina, V. (2020) Bioinformatics of nanopore sequencing. J. Hum. Genet., 65, 61–67. DOI
- Wick, R. R., Judd, L. M., Holt, K. E. (2019) Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol, 20, 129(2019). DOI
- Lanfear, R., Schalamun, M., Kainer, D., Wang, W., Schwessinger, B. (2019) MinIONQC: Fast and simple quality control for MinION sequencing data. Bioinformatics, 35(3), 523–525. DOI
- Li, H. (2018) Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics, 34(18), 3094–3100. DOI
- Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., Durbin, R. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25(16), 2078–2079. DOI
- Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., Kingsford, C. (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods, 14(4), 417–419. DOI
- Soneson, C., Yao, Y., Bratus-Neuenschwander, A., Patrignani, A., Robinson, M. D., Hussain, S. (2019) A comprehensive examination of Nanopore native RNA sequencing for characterization of complex transcriptomes. Nat Commun, 10, 3359(2019). DOI
- Workman, R. E., Tang, A. D., Tang, P. S., Jain, M., Tyson, J. R., Razaghi, R. et al. (2019) Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat Methods, 16(12), 1297–1305. DOI
- Zhang, P., Hung, L. H., Lloyd, W., Yeung, K. Y. (2018) Hot-starting software containers for STAR aligner. Gigascience, 7(8), giy092. DOI
- Pratt, B., Howbert, J. J., Tasman, N. I., Nilsson, E. J. (2012) Mr-Tandem: Parallel x!Tandem using Hadoop MapReduce on Amazon web services. Bioinformatics, 28(1), 136–137. DOI
- Data files produced by the GENCODE project. Retrieved September 1, 2020, from: ftp://ftp.ebi.ac.uk/pub/databases/gencode/_README.TXT
- Salmon Output File Formats. Retrieved September 1, 2020, from: https://salmon.readthedocs.io/en/latest/file_formats.html#fileformats