Effect of Geldanamycin Binding on the Thr90 Phosphorylation Site of Hsp90
Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 119121 Russia; *e-mail:veselov@ibmh.msk.su
Keywords:chaperone HSP90; prostate cancer; molecular dynamics; amino acid residue interaction network; RIN
DOI:10.18097/BMCRM00151
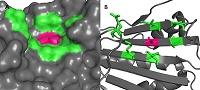
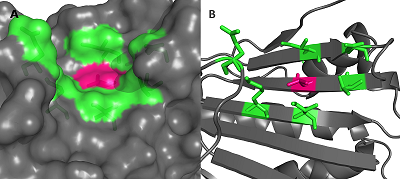
Prostate cancer is hormone-dependent and the androgen receptor (AR) is involved in its development. AR is a transcription factor that is activated by ligand binding, result in its translocation into the nucleus, where it initiates gene transcription. In an inactive state in cytoplasm AR exists as a complex with heat shock protein 90 (HSP90) and some other proteins. When the agonist binds, a conformational change in AR occurs, resulting in HSP90 and other chaperones dissociating. Recently it has been shown that for the dissociation of the HSP90-AR complex and the translocation of the latter into the nucleus, phosphorylation of the Thr-90 residue of the N-terminal domain of HSP90 is necessary. In this work, the effect of the HSP90 inhibitor, geldanamcin, interacting with the ATP-binding site, on the Thr90 phosphorylation site was investigated by molecular modeling methods. It has been shown that inhibitor binding slightly affects the size and mobility of cavity around Thr90. It is suggested that inhibitor binding to HSP90 does not result in changing the protein structure and does not influence on protein phosphory-lftion, and partially explains low effectiveness of such type of drugs in the therapy of prostate cancer.
|
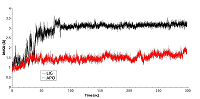
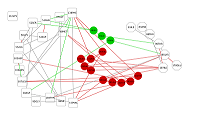
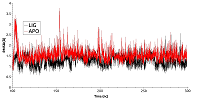
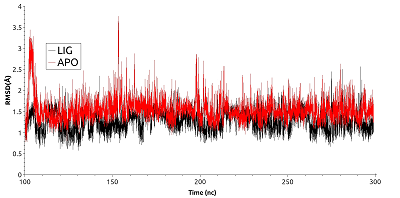
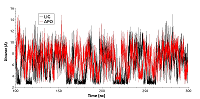
Figure 3.
RSMD for residues forming the cavity near phosphorylated Thr90 for free (red) and ligand bound (black) HSP90.
|
|
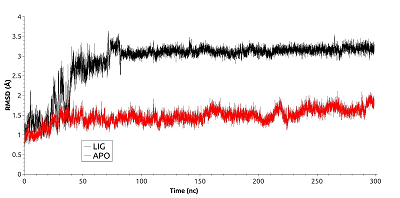
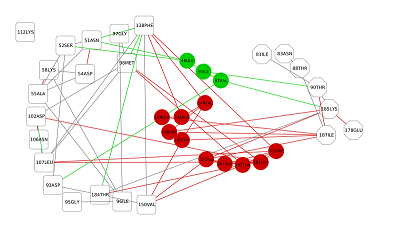
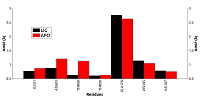
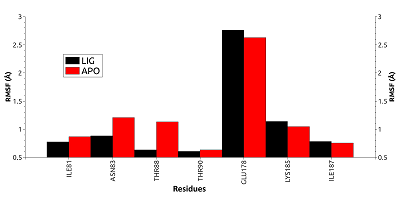
Figure 4.
RSMF for residues forming the cavity near phosphorylated Thr90 for free (red) and ligand bonded (black) HSP90.
|
|
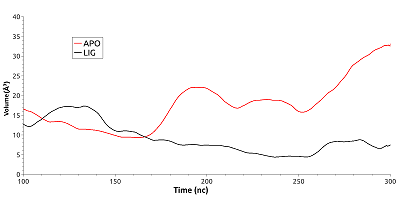
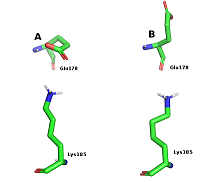
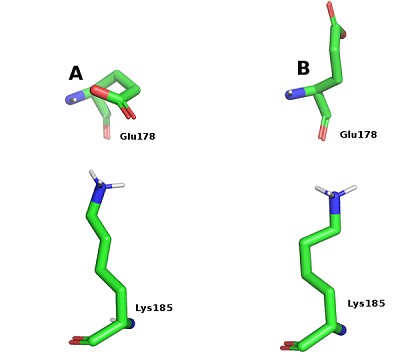
Figure 5.
Conformations of Glu178 and Lys185 during salt bridge formation (A) and Glu178 exposure into the solvent.
|
|
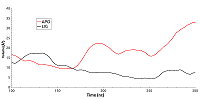
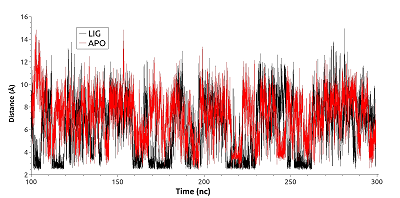
Figure 6.
The distance between centers of mass of amino group Lys185 and carboxyl group of Glu178 for free (red) and ligand bound (black) HSP90.
|
FUNDING
This work was supported by a grant from the Russian Foundation for Basic Research (project number 19-34-90057).
REFERENCES
- Culig, Z., Santer, F.R. (2014) Androgen receptor signaling in prostate cancer. Cancer Metastasis Rev., 33(2-3), 413-427. DOI
- Gelman, I.H. (2014) Androgen receptor activation in castration-recurrent prostate cancer: the role of Src-family and Ack1 tyrosine kinases. Int. J. Biol. Sci., 10(6), 620-626. DOI
- Culig, Z., Klocker, H., Bartsch, G., Hobisch, A. (2001) Androgen receptor mutations in carcinoma of the prostate: significance for endocrine therapy. Am. J. Pharmacogenomics, 1(4), 241-249. DOI
- Thakur, A., Roy, A., Ghosh, A., Chhabra, M., Banerjee, S. (2018) Abiraterone acetate in the treatment of prostate cancer. Biomed. Pharmacother., 101, 211-218. DOI
- Heinlein, C.A., Chang, C. (2004) Androgen receptor in prostate cancer. Endocr. Rev., 25(2), 276-308. DOI
- Dagar, M., Singh, J.P., Dagar, G., Tyagi, R.K., Bagchi, G. (2019) Phosphorylation of HSP90 by protein kinase A is essential for the nuclear translocation of androgen receptor. J. Biol. Chem., 294(22), 8699-8710. DOI
- Hoter, A., El-Sabban, M.E., Naim, H.Y. (2018) The HSP90 Family: Structure, Regulation, Function, and Implications in Health and Disease. Int. J. Mol. Sci., 19(9), 2560. DOI
- Solit, D.B., Scher, H.I., Rosen, N. (2003) Hsp90 as a therapeutic target in prostate cancer. Semin. Oncol., 30, 709-716. DOI
- Bohush, A., Bieganowski, P., Filipek, A. (2019) Hsp90 and Its Co-Chaperones in Neurodegenerative Diseases. Int. J. Mol. Sci., 20(20), 4976. DOI
- Wang, Y., McAlpine, S.R. Heat-shock protein 90 inhibitors: will they ever succeed as chemotherapeutics? (2015) Future Med. Chem., 7(2), 87-90. DOI
- Altieri, D.C. (2010) Mitochondrial Hsp90 chaperones as novel molecular targets in prostate cancer. Future Oncol., 6(4), 487-489. DOI
- Wei, T., Chang, C., Xue, L., Jieling, Zh., Jie, L. (2018) CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Research, 46, W363–W367. DOI
- Abraham, M.J., van der Spoel, D., Lindahl, E., Hess, B. and the GROMACS development team, (2020) GROMACS User Manual Release 2020
- Humphrey, W., Dalke, A., Schulten, K. (1996) VMD: Visual molecular dynamics. J. Mol. Graph., 14(1), 33–38. DOI
- The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC. (pymol.org/2/)
- Piovesan, D., Minervini, G., Tosatto, S.C.E. (2016) The RING 2.0 web server for high quality residue interaction networks. Nucleic Acids Research, 44(W1), W367-74. DOI
- Goenawan, I.H., Kenneth B., Lynn D.J. (2016) DyNet: visualization and analysis of dynamic molecular interaction networks. Bioinformatics, 32(17):2713-5. DOI
- Scardoni, G., Tosadori, G., Pratap, S., Spoto, F., Laudanna, C. (2015) Finding the shortest path with PesCa: a tool for network reconstruction. F1000Res., 4, 484. DOI
- Le Guilloux, V., Schmidtke, P., Tuffery, P. (2009) Fpocket: An open source platform for ligand pocket detection. BMC Bioinformatics, 10, 168. DOI
- Stebbins, C.E., Russo, A.A., Schneider, C., Rosen, N., Hartl, F.U., Pavletich, N.P. (1997) Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell, 89(2), 239-250. DOI