|
Methods for the Preparation of Blood Serum Samples in the Assessment of the Pharmacokinetics of Protein Drugs by the Example of Viscumin
1State Research and Testing Institute of Military Medicine, 4 Lesoparkovaya str., Saint-Petersburg, 195043 Russia; *е-mail: gniiivm_7@mil.ru 2Scientific center “Signal”, 16A Nagatinskaya str., Mosсow, 115487 Russia Key words: mass-spectrometry; serum; pharmacokinetics; lectins; quantitative analysis DOI: 10.18097/BMCRM00152 INTRODUCTION
The creation of new drugs includes mandatory trials of the safety and efficacy of pharmacotherapy. This is facilitated by the results obtained in the course of clinical studies of pharmacokinetics; the latter requires the development of a method for determining the components of drugs in biological material. The main research objects for the assessment of pharmacokinetic processes, such as distribution, absorption, elimination, are blood and urine samples. Nevertheless, whole blood is practically not used for determination of the content of physiologically active compounds of a protein nature, because blood components contain a large amount of related compounds that affect the sensitivity of the analysis method and reduce the recovery of target compounds. Clinical trials of medicinal products containing protein compounds, as a rule, involve the analysis of serum and plasma samples. Urine samples are not sufficiently informative for studying the pharmacokinetic parameters of protein compounds. In this regard, blood serum is the most representative test for assessing the pharmacokinetic parameters of medicinal compounds of a protein nature [1]. Blood serum is a complex matrix due to the presence of a large number of various proteins, complicating determination of target proteins. Currently, determination of protein compounds in biological material is carried out mainly using immunochemical methods, but they have a number of limitations. As a rule, preparation of samples of biological material is carried out by immunochemical methods and kits for isolation using specific antibodies, which have a limited shelf life. Immunochemical methods do not always have a sufficiently pronounced specificity; they are characterized by the risk of obtaining both false positive results due to cross reactions in the presence of related proteins, and false negative results in the presence of concomitant compounds in the matrix. In addition, there are objective difficulties in the production and purification of specific antibodies in relation to the entire variety of protein preparations. At the present stage of the development of analytical chemistry, the determination of exogenous proteins in blood serum without the use of antibodies is possible only if a particularly sensitive method of analysis is used, in particular, high-performance liquid chromatography with high-resolution mass spectrometric detection (HPLC-HRMS). In this connection, an urgent task is the development of alternative methods for the isolation of exogenous proteins at the stage of preparation of blood serum samples. Currently, promising and actively studied active components of antiblastoma and immunomodulatory drugs of the protein structure are mistletoe plant lectins, which exhibit marked antitumor and immunostimulating effect and have a slightly larger area of therapeutic application and less toxicity to cells in comparison with other lectins, from for example, castor bean (Ricinus Communis ), abrus (Abrus Precatorius), momordica (Momordica Balsaminum) [2-5]. This study on the development of approaches to the isolation of exogenous proteins from blood serum without the use of specific antibodies was carried out using mistletoe lectin (Viscum Album) - viscumin isoform MLI [6-7]. MATERIALS AND METHODS
Materials and Reagents The study of methods for preparing blood serum samples of laboratory animals (rats) was carried out using a sample of Viscum Album lectin, MLI visumin (Sigma Aldrich, Canada). Isolation of viscumin from a blood serum sample was carried out using the following methods: precipitation of proteins with 1% trichloroacetic acid in isopropanol (TCA/TPA) (Sigma Aldrich, Canada), purification from the fraction of the most pronounced proteins using ProteoMiner columns (Bio-Rad, USA), removal of albumin using the Albumin Depletion Kit (Thermo Scientific, USA) and Aurum Affi-Gel Blue columns (Bio-Rad). Enzymatic digestion of proteins of purified samples was carried out on centrifugal filters Amicon (Millipore, Ireland), volume 0.5 ml, having a membrane with a cut-off by weight of 10 kDa. In the process of developing a method for preparing samples, we used a Vanquish chromatographic system (Thermo Scientific) with a high-resolution hybrid mass-spectrometric detector Q Exactive HF-X, equipped with a quadrupole mass filter, a higher-energy C-trap dissociation (HCD) cell and an orbital ion trap (Thermo Scientific). Chromatographic separation of the components of the analyzed samples was carried out using a reverse phase C-18, Zorbax 300 SB analytical column (100 mm, i.d. 2.1 mm, particle size 3.5 μm) (Agilent, USA). A gradient elution mode was created using mobile phases: A - 0.1% formic acid (FA) (Fluka, USA) solution, and B - acetonitrile acetonitrile (ACN) (Sigma Aldrich, USA) with 0.1% formic acid.
Analysis Methods Evaluation of the Effectiveness of Methods for Extracting Viscumin from Blood Serum The choice of methods for the isolation of viscumin from blood serum samples without the use of specific antibodies was carried out on the basis of literature data [8-10], on non-immunochemical determination of protein compounds are presented. Model samples of viscumin were prepared according to the methods presented by Lee et al. [11]. Selection of a Solution for Solubilizing Precipitated Proteins The choice of the most effective reagent was carried out using the following solutions: 0.1% solution of the reagent Invitrosol (Inv), 0.1% solution of the reagent ProteaseMax Surfactant (PM) (Promega, USA), 0.1% solution of the RapiGest (RG) reagent (Waters, USA), 0.1% solution of the PPS Silent Surfactant (PPS) reagent (Agilent, USA). Additionally, two types of buffer s were used for solubilisation: 0.1% Tween-20 (Panreac, Spain) solution and 0.1 M Tris buffer solution, containing 8 M urea (Panreac) with addition of 0.1% Tween-20 (DB) to the final solution. After adding a 1% TCA/TPA to aliquots of blood serum, the precipitate containing the protein fraction was redissolved in the indicated reagent solutions and centrifuged at 12000 g and thermostating at 18°C. The supernatant was subjected to enzymatic digestion on a filter. Protein Precipitation with 1% TCA/IPA Precipitation was carried out using a 0.2 ml aliquot of blood serum by adding 2 ml of a 1% TCA/IPA. The resulting protein precipitate was washed with 0.2 ml of methanol, followed by removal of the supernatant layer. Then the precipitate was dissolved in 0.1 ml of 0.1% Tween-20. The aliquot was completely transferred to a centrifuge filter and subjected to enzymatic digestion. Removal of Matrix Proteins Using the ProteoMiner Kit (Bio-Rad) The purification of blood serum using ProteoMiner Large Capacity columns was carried out by taking 1 ml of the sample with the addition of viscumin in the dilution range (0.5–100) μg/ml. The procedure was carried out in accordance with the recommendations of the column manufacturer [12]. The purified fraction was transferred to a filter and subjected to enzymatic digestion. Albumin Removal Using the Albumin Depletion Kit (Thermo Scientific) Removal of albumin from blood serum by means of the Albumin Depletion Kit was performed using a 0.03 ml sample with addition of visumin in the dilution range (0.5–100) μg/ml. The serum sample was purified in accordance with the recommendations of the kit manufacturer [13]. The fraction purified from albumin was transferred to a centrifuge filter and subjected to enzymatic digestion. Removal of Albumin with the Aurum Affi-Gel Blue Kit (Bio-Rad) The preparation was carried out using a 0.125 ml sample with the addition of viscumin in the dilution range (0.5–100) µg/ml. The serum sample was purified in accordance with the recommendations of the column manufacturer [14]. The fraction purified from albumin was transferred to a centrifuge filter and subjected to enzymatic digestion. Enzymatic Cleavage of Proteins Protein preparation and subsequent proteolysis were performed by filter-aided sample preparation (FASP) suitable for the cleavage of proteins of cell or tissue lysates containing various detergents and widely used in proteomic studies [15-17]. The FASP method uses a filter with a molecular weight cut-off membrane as a "reactor", where complex protein mixtures are modified and hydrolyzed, and also effectively remove detergents and excess reagents that are not recommended for use in sample preparation for mass-spectrometric detection [18, 19]. The preparation of a sample containing viscumin for the determination of the HPLC-MS characteristics was carried out according to the FASP protocol proposed by Wiśniewski et al. [20] with some changes to increase the sensitivity and reproducibility of the method. In particular, the procedure for preparing samples for enzymatic digestion was carried out on centrifugal filters with a 10 kDa cutoff. The filters were preliminarily kept for 16 h in a 0.1% of Tween-20 to reduce the sorption of protein compounds and peptides on the filter membrane. 4-vinylpyridine (Sigma Aldrich) was used as an alkylating agent, it is more stable to oxidation, and forms stronger derivatives with cysteine. This makes it possible to reduce the number of gaps in the determination of the amino acid sequence (AAS) of proteins during the analysis of mass spectra of peptides. The filter was washed between the stages of denaturation, reduction, and alkylation with a buffer without urea: 0.1 M Tris, pH 8.5. The buffer was changed before proteolysis by washing the filter twice with 0.1 ml of buffer 0.05 M Tris, pH 8.0, followed by centrifugation with acceleration 12000 g and thermostating at 18°С. The filter was moved to a new sample receiver and 0.1 ml of buffer 0.05 M Tris, pH 8.0 was added to the filter. Enzymatic digestion of proteins usually is carried out with trypsin (cleaving peptide bonds formed by carboxyl groups of R and K), which makes it possible to obtain peptides with molecular weights from 400 Da to 2000 Da - the optimal mass range for analysis by mass-spectrometry [21]. In the presented study, a mixture of trypsin and lysC (cleaved proteins after the K and P site, except for sites КP, RP) was used to obtain a more informative peptide map. Proteins were cleaved with a mixture of trypsin and lysC enzymes (Promega) in 0.1 M HCl with a total concentration of 0.2 mg/ml in a volume of 0.01 ml for 16 h at 37°С. After proteolysis, the filter was centrifuged at 12000 g and thermostating at 18°С until the solution was completely removed from the filter. The final elution of peptides from the filter was carried out by adding 0.05 ml of a 0.1% FA (v/v) in a solution of ACN and deionized water in a ratio of 20:80 % (v/v) to the filter and centrifuged to dryness with acceleration 12000 g for 15 min at 18°С. The procedure was performed twice. The resulting mixture of peptides in the sample receiver was dried using a vacuum concentrator at 60°C to a minimum volume (0.005 ml). The peptide concentrate was then diluted in 0.05 ml of 0.1% (v/v) FA in a solution of ACN and deionized water in a ratio of 2:98 % (v/v), vortexed and centrifuged. The final peptide solution was transferred to a 0.25 ml silanized vial insert and analyzed by HPLC-HRMS. Conditions for Mass-Spectrometric Detection The analysis of peptides obtained during the enzymatic digestion of a 0.01 mg/ml viscumin sample was carried out by mass spectrometric detection with peptide ionization by electrospray in the positive ionization mode (ESI+). The registration of peptides was carried out in two sequentially switching modes of the mass spectrometer: - Full Scan MS (Full Scan) - scanning of the total ionic current of precursor ions in a given range of m/z values; - Data Depended MS2 (dd-MS2) - mass spectrometric detection method based on scanning the total ion current of fragment ions obtained during the dissociation of the 10 most intense precursor ions. The determination of viscumin peptides was carried out by analyzing the obtained mass chromatograms using the PeakStudio X software package from Bioinformatics Solutions Inc. (Canada). The parameters for processing mass spectra included the following established limitations: the permissible mass error tolerance for detection of product ions was 0.02 Da, the maximum permissible amount of simultaneously present post-translational modifications (PTMs) in a peptide was 3 from 4 possible: carbamidomethylation (constant modification), S-pyridylethylation (C), deamidation (N, Q) and oxidation (M) (variable modifications), and the permissible precursor mass error tolerance was 15 ppm. Further targeted analysis by HPLC-HRMS with detection in two sequentially switching modes of the mass spectrometer: Full Scan and Parallel Reaction Monitoring (PRM). The PRM mode is characterized by a higher sensitivity compared to dd-MS2 mode due to the isolation in the first quadrupole of the target ions of the peptides specified in the method parameters, their further fragmentation in the collision cell and detection of product ions in the Orbitrap mass analyzer. Protein identification was carried out according to the four selected viscumin peptides. The quantitative assessment of the lectin content in the model blood serum samples was carried out using one of the most intense peptide ion. Conditions for Chromatographic Separation of Peptides in Analysis by Analysis-Dependent Mass Spectrometric Scanning Chromatographic separation of a mixture of peptides was carried out in the gradient elution mode: from 0 to 2 min 2% mobile phase B, from 2 min to 38 min linear gradient to 45% mobile phase B, from 38 min to 39 min linear gradient to 95% mobile phase B, from 39 min to 44 min 95% mobile phase B, 44 min to 45 min linear gradient up to 2% mobile phase B, up to 50 min 2% mobile phase B. Conditions for Chromatographic Separation of Peptides in Analysis by Monitoring Parallel Reactions Gradient elution of peptides was performed as follows: from 0 to 1 min 2% mobile phase B, from 1 min to 12 min linear gradient to 40% mobile phase B, from 12 min to 13 min linear gradient to 95% mobile phase B, from 13 min to 16 min 95% mobile phase B, from 16 min to 16.01 min linear gradient up to 2% mobile phase B, 2% mobile phase B up to 35 min. RESULTS In the process of developing a method for preparing samples for quantitative analysis of the viscumin content in blood serum, studies were carried out to select characteristic peptides, i.e. present only in the composition of a particular protein compound. Based on the results of the analysis of a sample of the viscumin peptide lysate (0.01 mg/ml), carried out by the DDA method of mass-spectrometric scanning, lectin peptides were identified and the most intense characteristic peptides were selected: two- or three-charged molecular ions with the highest signal intensity per mass chromatogram without modified amino acid residues (Table 1).
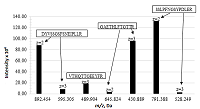
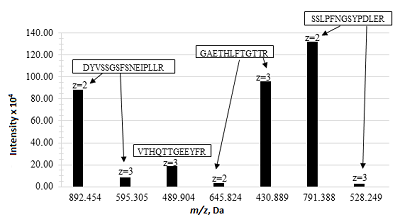
Analysis of the data obtained made it possible to identify the following most intense peptide ions: DYVSSGSFSNEIPLLR (m/z 892.454, z = 2+), GAETHLFTGTTR (m/z 430.889, z = 3+), and SSLPFNGSYPDLER (m/z 791.388, z = 2+). The peptide SSLPFNGSYPDLER (m/z 791.388, z = 2+), which has the maximum intensity and meets all the requirements for characteristic peptides, was chosen as the peptide ion for quantitative assessment (Fig. 1).
The selected peptides were used to develop the PRM method, which could shorten the analysis time and increase the sensitivity in the determination of viscumin. Precipitation with organic solvents leads to denaturation of protein molecules, as a result of which their solubility deteriorates. When using the precipitation of proteins with 1% TCA/IPA, the solubility of the precipitated proteins in solutions of various types of surfactants was additionally evaluated. As a result of evaluating the most effective solution for the reconstitution of proteins precipitated with 1% TCA/IPA, a 0.1% Tween-20 was selected (Table 2) with the degree of viscumin extraction from blood serum (8.2 ± 2.2)%.
The results of determining the content of viscumin in model blood serum samples with the addition of lectin in the concentration range (0.5–100) μg/ml, prepared by various methods, are presented in Table 3.
DISCUSSION Evaluation of sample preparation methods according to the minimum detectable concentration has shown that of among the presented methods for purifying blood serum (Supplementary materials), the most effective is the method using ProteoMiner spin columns, it allows lectin quantitative determination in serum at a level of 0.5 μg/ml with a degree of extraction (2.7 ± 0.6)%. The highest degree of extraction of lectin (6.9 ± 1.6)% was achieved by the method of protein precipitation with 1% TCA/IPA, but with the lowest sensitivity (5 μg/ml). The principle of operation of the ProteoMiner columns is based on the difference in the affinity of blood serum components to ligands bound to the column sorbent, which made it possible to isolate the target protein in the lowest concentration. Sample preparation by the described methods was carried out for different initial volumes of serum samples, which was not taken into account in the aboveThe results of determining the content of viscumin in model blood serum samples with the addition of lectin in the concentration range (0.5–100) μg/ml, prepared by various methods, are presented in Table 3. For this reason, it is advisable to compare the effectiveness of the approaches chosen at the first stage for equal volumes of the initial sample. A comparative study of methods for preparing blood serum was carried out using different initial volumes for purification of model samples: 0.2 ml, 0.5 ml, 1.0 ml with the introduction of the following concentrations of viscumin: 1 μg/ml, 5 μg/ml, 10 μg/ml. Sample preparation was carried out by removing albumin by precipitation in 1% TCA/IPA and purification on ProteoMiner spin columns.
The results of the study, presented in Table 4, showed that an increase in the sample volume did not lead to an increase in the degree of extraction of viscumin from the blood serum. The use of ProteoMiner columns and the method for removing albumin by precipitation in a 1% TCA/IPA solution are characterized by an equivalent level of viscumin release with a recovery rate of within 7% (in a sample volume not exceeding 0.2 ml). However, the detection limit of lectin in the case of using the 1% TCA/IPA for albumin removal is significantly reduced with a decrease in its concentration in serum. Thus, according to the results of the study, it was found that when using ProteoMiner columns, the sensitivity of the method for preparing blood serum by removing major proteins exceeded 0.5 μg/ml. This is significantly higher than when using the precipitation method with 1% TCA/IPA. CONCLUSIONS Approaches without the use of specific antibodies can be used as alternatives to immunochemical methods for preparing blood serum samples for the isolation and quantitative assessment of the content of protein components of drugs. Using the example of a component of the medicinal product of the lectin of the plant Viscum Album, viscumin MLI, the limits of its determination in serum are established. As a result of a comparative assessment, the optimal method for preparing blood serum samples using ProteoMiner columns was selected. The proposed method, together with the HPLC-MS method, is recommended to be used for quantitative assessment of the content of target components during pharmacokinetic studies of drugs in cases where the use of specific antibodies for one reason or another is not possible. COMPLIANCE WITH ETHICAL STANDARDS The work does not contain research using people as research objects. All applicable international, national and/or institutional guidelines for the care and use of animals have been followed. ACKNOWLEDGMENTS The authors are grateful to the management of the State Research and Testing Institute of Military Medicine for the opportunity to conduct this research work. FUNDING The study was carried out on the topic of research work in the framework of the implementation of the State Defense Order. CONFLICT OF INTEREST The authors declare no conflicts of interest. Supplementary materials are available at http://dx.doi.org/10.18097/BMCRM00152. REFERENCES
|