Algorithms for Calculation of Parameters of Electrochemical Biosensor
1Institute of Biomedical Chemistry, Pogodinskaya Street, 10, Moscow 119121, Russia; *e-mail: viktoria.shumyantseva@ibmc.msk.ru
2Pirogov Russian National Research Medical University, Moscow, Russia
Keywords: electrochemical biosensor; electroactive electrode surface; binding constant; DNA analysis; pharmacogenomics
DOI:10.18097/BMCRM00178
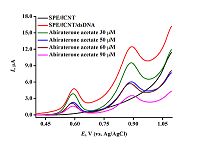
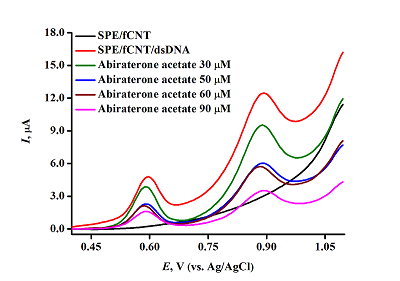
The aim of this work is to present the experimental results in the form of an algorithm for analyzing the modification of screen printed electrodes, including the possibility of its regeneration for irreversibly oxidizing biologically active compounds (drugs, DNA and proteins). A protocol was developed for quantitative analysis and study of the mechanism of drug-DNA interaction by differential pulse voltammetry, including the following parameters: complex binding constant, Gibbs free energy, and electrochemical coefficients of the toxic effect.
|
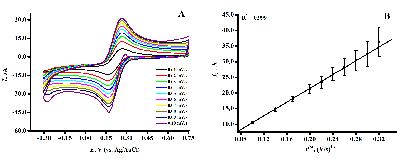
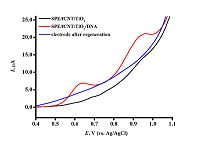
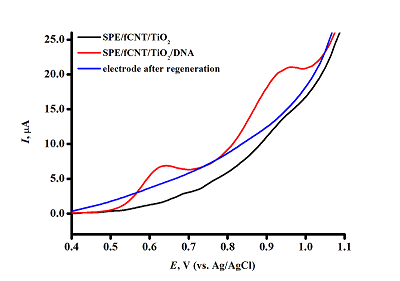
Figure 5.
DVPs SPE/fCNT/TiO2 with dsDNA concentration of 3 mg/ml. (SPE – screen-printed electrode, fCNT – functionalized carbon nanotubes).
|
|
CLOSE

|
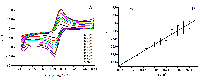
Table 1.
Electroanalytical characteristics of screen printed electrodes (SPE) modified with fCNT/TiO2 obtained in a 5 mM solution of potassium hexacyanoferrate. The potential values and peaks of oxidation and reduction of potassium ferricyanide are presented for a potential sweep rate of 50 mV/s. (fCNT – functionalized carbon nanotubes).
|
|
CLOSE

|
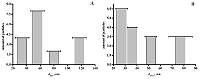
Table 2.
Electroactive areas of SPE modified with various dispersions ((SPE – screen-printed electrode, fCNT – functionalized carbon nanotubes).
|
|
CLOSE

|
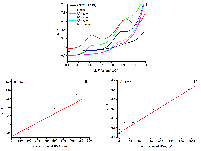
Table 3.
Metrological and electrochemical parameters of the electrodes.
|
FUNDING
The work was performed within the framework of the Program for Basic Research in the Russian Federation for a long-term period (2021-2030) (№122030100168-2).
REFERENCES
- Mostafa M.I., Tian Y., Anjum S., Hanif S., Hosseini M., Lou B., Xu G. (2022) Comprehensive review on the electrochemical biosensors of different breast cancer biomarkers. Sensors and Actuators B: Chemical, 365, 131944. DOI
- Pepe M.S., Etzioni R., Feng Z., Potter J. D., Thompson M.L., Thornquist M., Winget M., Yasui Y. (2001) Phases of Biomarker Development for Early Detection of Cancer. JNCI: Journal of the National Cancer Institute, 93, 14, 1054–1061. DOI
- 3. Lu D., Xu Q., G. Pang G. (2019) A bombykol electrochemical receptor sensor and its kinetics. Bioelectrochemistry, 128, 263-273, 1567-5394. DOI
- Ronkainen N.J., Halsall H.B., HeinemanW.R. (2010) Electrochemical biosensors. Chem. Soc. Rev., 39, 1747–1763. DOI
- Ghosh M., Mandal S., Roy A., Mondal P., Mukhopadhyay S. K., Chakrabarty S., Chakrabarti G., Pradhan S.K. (2021) Synthesis and characterization of a novel nanocarrier for biocompatible targeting of an antibacterial therapeutic agent with enhanced activity. Journal of Drug Delivery Science and Technology, 66, 102821. DOI
- Randles J.E.B. (1948) A cathode-ray polarograph. Part II - The current-voltage curves. Trans Faraday Soc., 44, 327.
- Sevcik A. (1948) Oscillographic polarography with periodical triangular voltage. Collect Czech ChemCommun, 13, 349.
- Mohammadi A., Moghaddam A.B., Alikhani E., Eilkhanizadeh K., Mozaffari S. (2013) Electrochemical quantification of fluoxetine in pharmaceutical formulation using carbon nanoparticles. Micro & Nano Letters, 8, 853-857. DOI
- Ferapontova E.E. (2018) DNA Electrochemistry and Electrochemical Sensors for Nucleic Acids. Annual review of analytical chemistry, 11 (1), 197–218. DOI
- Kuzikov, A.V., Bulko, T.V., Koroleva, P.I., Masamrekh, R.A., Babkina, S.S., Gilep, A.A., Shumyantseva, V.V. (2020) Cytochrome P450 3A4 as enzyme for drug biotransformation: the role of sensor systems modifications in electrocatalysis and electroanalysis. Biomeditsinskaya Khimiya, 66 (1), 64-70. DOI
- Shumyantseva, V.V., Sigolaeva, L.V., Agafonova, L.E., Bulko, T.V., Pergushov, D.V., Schacher, F.H., Archakov, A.I. (2015) Facilitated biosensing via direct electron transfer of myoglobin integrated into diblock copolymer/multi-walled carbon nanotube nanocomposites. J. Mater. Chem. B, 3(27) 5467–5477. DOI
- Shumkov, A. A., Suprun, E. V., Shatinina, S. Z., Lisitsa, A. V., Shumyantseva, V. V., Archakov, A. I. (2013) Gold and Silver Nanoparticles for Electrochemical Detection of Cardiac Troponin I based on Striping Voltammetry. BioNanoScience, 2 (3), 216-222. DOI
- Nimal R., Unal D. N., Erkmen C., Bozal-Palabiyik B., Siddiq M., Eren G., Shah A., Uslu B. (2022) Development of the electrochemical, spectroscopic and molecular docking approaches toward the investigation of interaction between DNA and anti-leukemic drug azacytidine. Bioelectrochemistry, 146, 108135,1567-5394. DOI
- Topkaya S.N., Kaya H.O., Cetin A.E. (2021) Electrochemical detection of linagliptin and itsinteraction with dna. Turkish J. Pharm. Sci., 18, 645–651. 24 DOI
- Manzano M., Viezzi S., Mazerat S., Marks R.S., Vidic J. (2018) Rapid and label-free electrochemical DNA biosensor for detecting hepatitis A virus. Biosens. Bioelectron, 100, 89–95. DOI
- Lavín, Á., Vicente, J.D., Holgado, M., Laguna, M.F., Casquel, R., Santamaria, B., Maigler,M.V., Hernandez, A.L., Ramirez, Y. (2018) On the Determination of Uncertainty and Limit of Detection in Label-Free Biosensors. Sensors, 18, 2038. DOI
- Armbruster, D.A., Pry, T. (2008) Limit of blank, limit of detection and limit of quantitation. Clin.Biochem. Rev., 29, S49–S52.
- Bolat G. (2020) Investigation of poly(CTAB-MWCNTs) composite based electrochemical DNA biosensor and interaction study with anticancer drug Irinotecan. Microchemical Journal, 159, 105426. DOI
- Hasanzadeh M., Shadjou N. (2016) Pharmacogenomic Study Using Bio- and Nanobioelectrochemistry: Drug–DNA Interaction. Mater. Sci. Eng. C, 61, 1002–1017. DOI
- Acharya, M., Bernard, A., Gonzalez, M., Jiao, J., De Vries, R., Tran, N. (2012) Open-label, phase I, pharmacokinetic studies of abiraterone acetate in healthy men. Chemother.Pharmacol., 69, 1583−1590. DOI
- Gurova K., (2009) New hopes from old drugs: revisiting DNA-binding small molecules as anticancer agents. Futur. Oncol.,5, 1685 DOI
- Muti M. (2018) Electrochemical monitoring of the interaction between anticancer drug and DNA in the presence of antioxidant. Talanta., 178, 1033–1039. DOI
- Bagni G., Osella D., Sturchio E., MasciniM. (2006) Deoxyribonucleic acid (DNA) biosensors for environmental risk assessment and drug studies. Anal.Chim.Acta.,81–89, 573-574. DOI
- Shumyantseva V.V., Bulko T.V., Tikhonova E.G., Sanzhakov M.A., Kuzikov A.V., Masamrekh R.A., Pergushov D.V., Schacher F.H., Sigolaeva L.V. (2021) Electrochemical studies of the interaction of rifampicin and nanosome/rifampicin with dsDNA. Bioelectrochemistry, 140, 107736. DOI
- Wani T.A., Alsaif N., Bakheit A.H., Zargar S., Al-Mehizia A.A., KhanA.A. (2020) Interaction of an abiraterone with calf thymus DNA: Investigation with spectroscopic technique and modelling studies. Bioorg Chem., 100, 103957. DOI
- Nafisi S., Saboury A.A., Keramat N., Neault J.F., Tajmir-RiahiH.A. (2007) Stability and structural features of DNA intercalation with ethidium bromide, acridine orange and methylene blue. J. Mol. Struct.,827, 35–43. DOI
- Sirajuddin M., Ali S., BadshahA. (2013) Drug–DNA interactions and their study by UV–vis, fluorescence spectroscopies and cyclic voltammetry. J. Photochem. Photobiol. B: Biol.,124, 1–19. DOI
- DeDogan-Topal B., Bozal-Palabiyik B., Ozkan S.A., UsluB. (2014) Investigation of anticancer drug lapatinib and its interaction with dsDNA by electrochemical and spectroscopic techniques. Sens. Actuators B Chem.,194 185–194. DOI
- Temerk Y., Ibrahim M., Ibrahim H., KotbM. (2016) Interactions of an anticancer drug lomustine with single and double stranded DNA at physiological conditions analysed by electrochemical and spectroscopic methods. J. Electroanal. Chem.,769, 62–71. DOI
- Yazan Z., BayraktepeD.E., Dinç.E. (2020) Four-way parallel factor analysis of voltammetric four-way dataset for monitoring the etoposide-DNA interaction with its binding constant determination.Bioelectrochemistry, 134, 107525. DOI
- Chaires J.B., (2006) A thermodynamic signature for drug-DNA binding mode., Arch. Biochem.Biophys., 453, 26–31. DOI