Algorithm of targeted (SRM) methods development in mass-spectrometric studies
Institute of Biomedical Chemistry, 119121 Moscow, Pogodinskaya Str., 10 bldg. 8, *e-mail: victor.zgoda@gmail.com
Keywords: selected reaction monitoring; peptides; stable isotope labeling; mass spectrometry
DOI: 10.18097/BMCRM00006
Determination of protein concentration in biological samples is an important task for biological research, as well as for medicine and routine clinical biochemistry.
The introduction of stable isotope-labeled peptide standards (SIS) made it possible to determine accurately absolute protein concentrations in proteomic studies. The correct choice of SIS and the systematic way to develop a method for selected reaction monitoring (SRM) are very important steps that are crucial for further identification and measurements of protein concentration. In this paper, we summarize our experience of selecting SIS for measuring the protein concentration by SRM. The results are presented in the form of an algorithm that describes the main stages of the SIS selection and the main points in the development of SRM methods for the targeted protein detection and determination of protein concentrations in biological samples.
|
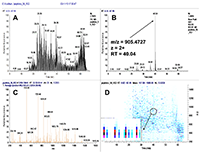
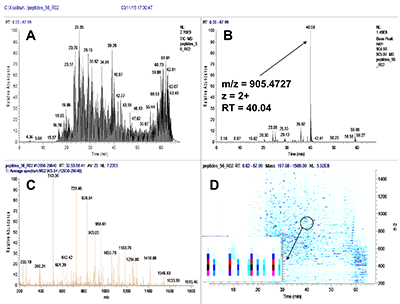
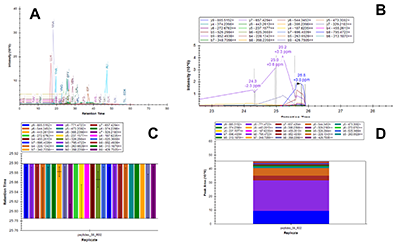
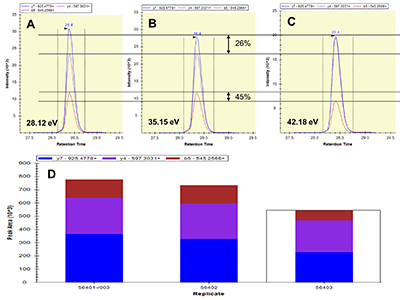
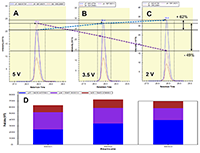
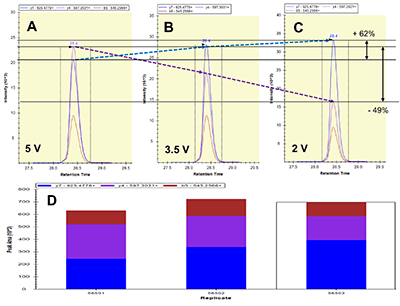
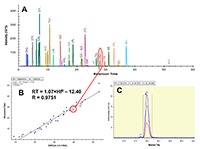
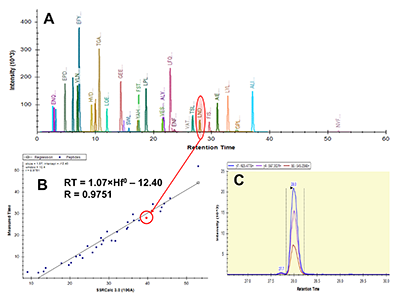
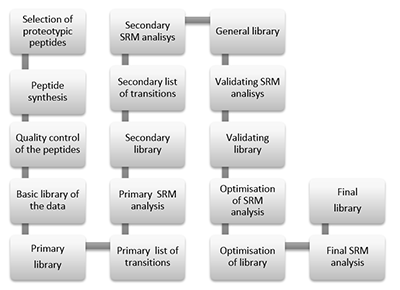
Figure 1.
General steps of peptide selection and SRM method development for proteins quantitation by SIS.
|
ACKNOWLEDGEMENTS
The work was supported under the auspice of the Russian Academy of Sciences research program "Fundamental Research for Biomedical Technologies" for 2018.
SUPPLEMENTARY
REFERENCES
- Aebersold, R., & Mann, M. (2016). Mass-spectrometric exploration of proteome structure and function. Nature, 537(7620), 347–355. DOI
- Chambers, A. G., Percy, A. J., Simon, R., & Borchers, C. H. (2014). MRM for the verification of cancer biomarker proteins: recent applications to human plasma and serum. Expert Review of Proteomics, 11(2), 137–148. DOI
- Wu, H.-Y., Goan, Y.-G., Chang, Y.-H., Yang, Y.-F., Chang, H.-J., Cheng, P.-N., et al. (2015). Qualification and Verification of Serological Biomarker Candidates for Lung Adenocarcinoma by Targeted Mass Spectrometry. Journal of Proteome Research, 14(8), 3039–3050. DOI
- Percy, A. J., Tamura-Wells, J., Albar, J. P., Aloria, K., Amirkhani, A., Araujo, G. D. T., et al. (2015). Inter-laboratory evaluation of instrument platforms and experimental workflows for quantitative accuracy and reproducibility assessment. EuPA Open Proteomics, 8, 6–15. DOI
- Hood, C. A., Fuentes, G., Patel, H., Page, K., Menakuru, M., & Park, J. H. (2008). Fast conventional Fmoc solid-phase peptide synthesis with HCTU. Journal of Peptide Science?: An Official Publication of the European Peptide Society, 14(1), 97–101. DOI
- Moskaleva, N., Moysa, A., Novikova, S., Tikhonova, O., Zgoda, V., & Archakov, A. (2015). Spaceflight Effects on Cytochrome P450 Content in Mouse Liver. PloS One, 10(11), e0142374. DOI