|
In Silico Analysis of Interaction between Seaweed-Derived Bioactive Compounds and Selected Diabetes-Related Targets
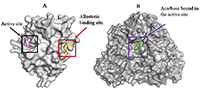
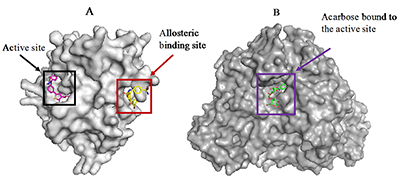
1Centre for Bio-computing and Drug Development, Adekunle Ajasin University, Akungba-Akoko, Key words: protein tyrosine phosphatase 1B; seaweed components; diabetes, docking; α-glucosidase DOI: 10.18097/BMCRM00074 INTRODUCTION Diabetes mellitus (DM) is a common metabolic disorder associated with chronic hyperglycemia occasioned by defects in insulin action and/or secretion resulting in imbalance of metabolism of proteins, carbohydrates, and lipids. Prolonged hyperglycemia may lead to other serious complications [1,2]. Although there is no cure for the disease till date, enomorous efforts have been undertaken to control blood glucose especially in DM type 2 patients as one of the strategies to manage the disease and to prevent the associated complications. The list of validated proteins targeted for inhibition in antidiabetic therapy includes protein tyrosine phosphatase (PTP) 1B and α-glucosidase. PTP1B is a member of the PTP superfamily and a negative regulator of multiple receptor tyrosine kinases [3]. Inhibition of PTP1B has been proposed as a strategy for the treatment of DM type 2 and obesity [4]. This protein regulates both insulin and leptin signaling pathways. Although, certain attempts have been made to develop potent and selective inhibitors targeting the active site of PTP1B, it is a difficult task due to high homogeneity of PTP1B to other cellular PTPs. In addition, the polar nature of the active site pocket is another challenge. However, available reports on the structure and enzyme kinetics of PTP1B have hinted the unique structural requirements for substrate and inhibitor recognition [5-7]. The recent discovery of a novel allosteric site recognized in the PTP1B crystal structure [7] has proved this site as an ideal binding pocket for small molecule drug discovery because, in contrast to the active site, the allosteric site is not well conserved among phosphatases [3,7]. Hence, it affords an opportunity to potentially bypass the challenges associated with catalytic site inhibition. On the other hands, α-glucosidase is an enzyme that plays a key role in carbohydrate degradation through hydrolysis of complex sugars into their corresponding monomers. The enzyme is the therapeutic target for acarbose [8], a potent carbohydrate mimetics currently used to control the postprandial blood glucose level in DM patients. The strategy is to inhibit the catalytic function of the enzyme to prevent catabolism of starches, disaccharides, and oligosaccharides [9]. Inhibitors from medicinal plants, microorganisms, and marine algae have been preferable for treatment of DM via α-glucosidase inhibition [10,11]. Marine seaweeds are reputed as a natural and rich source of bioactive ingredients over the years. The phytochemicals found in marine algae include polyphenols and phlorotannins. These phytoconstituents determine potent biological activities of seaweeds [12,13]. Other components like carotenoids, polysaccharide and glycoproteins found in the sea algae have been researched for specific bioactivities in animal models and in in vitro experiments [14-16]. Accordingly, seaweeds have long been applied in folkloric medicine as remedies for therapy of various diseases. Nutritionally, seaweeds contain vitamins and minerals, as well as micronutrients and macronutrients [17]. In this context anticancer, hepatoprotective, antioxidant, immunomodulatory, anti-coagulant, antidiabetic, and anti-hypercholesterolemic effects have been reported for marine algae and their components [18-22]. Much attention is paid to their therapeutic benefits in prevention as well as management of DM and its complications [23-26]. The brown algae, Eisenia bicyclis (E. bicyclis), Ecklonia stolonifera (E. stolonifera), Ecklonia maxima (E. maxima), and Ecklonia cava (E. cava), are well documented for their desirable pharmacological potentials in the management of DM. They are all locally consumed as foodstuff in different parts of the world. E. stolonifera («tutuarame») is a brown alga which is traditionally found in Asia especially in Japan [23-25]. The alga contains oxylipins and phlorotannins, the components that are responsible for pharmacological potentials of the plant. E. bicyclis and E. cava are abundantly distributed in the middle Pacific coast and southern coast of Japan and Korea respectively. E. bicyclis has been reported for numerous therapeutic effects like anti-microbial, anti-diabetic, anti-hyperlipidemic, algicidal, anticancer, tyrosinase inhibitory, anti-skin aging, anti-inflammatory, hepatoprotective, antioxidant, nitrite-scavenging, anti-plasmin inhibitory, anti-atherosclerosis, anti-Alzheimer’s disease, angiotensive converting enzyme- and anti-allergic activities [27-30]. Similarly, E. cava possesses many pharmacological effects such as anti-hypertensive, radioprotective, anti-allergic, anti-inflammatory, anti-proliferative, anti-diabetic, antioxidant, anti-human immunodeficiency virus (HIV) and hyaluronidase inhibitory activities [31-34]. E. maxima is peculiar to Africa continent, reportedly found in South Africa and Namibia where it is called «sea bamboo» and used for kelpak production. Researchers have claimed antioxidant and anti-diabetic properties for this marine alga [31,35]. Despite the growing acceptance of sea food consumption as an essential strategy in diabetes prevention and management, the mechanisms underlying their observed antidiabetic bioactivities especially at the molecular level remain to be fully unraveled. Furthermore, their precise interaction with diabetes-related proteins such as α-amylase, α-glucosidase, aldose reductase, peroxisome proliferator-activated receptor gamma and PTP1B has not been exhaustively investigated. Recently, interaction between aldose reductase and algae-derived compounds was studied used in silico analysis [36]. The current study is designed to depict the affinity of selected seaweed components and their molecular binding signatures for α-glucosidase and PTP1B as validated antidiabetic targets. The results obtained in this work are relevant in validating the folkloric use of these seaweeds in the management of DM.MATERIALS AND METHODS Virtual Preparation of Target Proteins Crystal structures of human PTP1B (PDB ID: 1T49) and α-glucosidase (3TOP) used in this study were retrieved from the Brookhaven protein data bank (http://www.rcsb.org/pdb). The macromolecules were visualized using the Python-enhanced molecular graphics tool (PyMol). PTP1B was found in the complex with «inhibitor 3», a potent allosteric inhibitor of PTP1B while α-glucosidase was co-crystallized with acarbose. Crystallographic water molecules which were found with structures were then deleted prior molecular docking procedure [37]. The putative binding sites of the proteins were identified with reference to the co-crystallized ligands. Ligands Selection, Preparation and Optimization Chemical structures of phlorofucofuroeckol-A, dieckol, eckol, dioxinodehydroeckol, phloroglucinol and 7-phloroeckol were retrieved from the NCBI PubChem database (http://www.ncbi.nlm.nih.gov/pccompound) in the 3D (.sdf) format. Acarbose, the reference ligand for α-glucosidase, and «inhibitor 3», the control ligand for PTP1B were also downloaded from NCBI PubChem database. Identification and selection of the seaweed components was based on information from the literature. Molecular Docking and Scoring For ligand docking and target-ligand complex analysis, Autodock Vina suite on PYMOL was used [38,39]. First, based on the already present co-crystallized ligand in the pdb file, the inhibitor binding site was defined with grid parameters set to include all the amino acid residues at the putative binding site. The spacing between grid points was maintained at 0.375 angstroms. This gives enough space to enhance adequate ligand rotation and translation. All ligands were docked to the putative binding pocket of the proteins. Throughout this experiment, the rotatable bonds of the ligands were set to be free while the protein molecule was treated as a rigid structure. Repeated docking runs were performed for each ligand with the number of modes set to 10 to achieve more accurate and reliable results. The best results obtained based on the binding configuration and binding affinity were chosen for further analysis. Validation of Molecular Docking Protocol Molecular docking validation was carried out as previously described [37]. Briefly, the ligand found at the binding site of the experimentally determined protein crystal was deleted. Then, the ligand (.sdf format) was separately prepared using Marvin sketch as described above and re-docked into the active site. The binding pose and molecular interaction pattern was compared to that of the x-ray diffraction crystal structure. Data Analysis The molecular graphics program PyMol intended for the structural visualization of proteins was used to obtain the protein-ligand snapshots while proteinsplus was used to depict details of protein-ligand interactions. Proteinsplus (http://proteinsplus.zbh.uni-hamburg.de/) is an online server which presents detailed protein-ligand interactions especially hydrogen bond, hydrophobic bond and pie-stacking interactions [40,41]. RESULTS AND DISCUSSION The search for natural compounds with potential antidiabetic activity is still encouraged because the disease still remains without effective cure till date. α-Glucosidase and PTP1B have evolved as promising targets in the management of DM. Since E. bicyclis, E. stolonifera, Ecklonia maxima, and E. cava have been valuably explored in the folkloric and traditional medicine for the treatment of obesity and diabetes, we employed computational techniques to unravel the precise molecular interaction of the algae-derived compounds with human PTP1B (Fig. 1a) and α-glucosidase (Fig. 1b) having clinical relevance in both diseases.
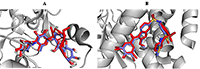
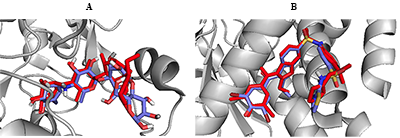
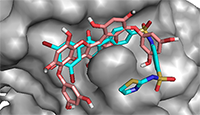
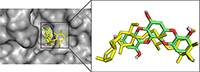
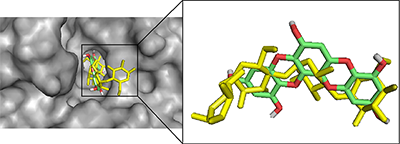
In order to validate the docking protocol towards ensuring accuracy and reliability of the protein-ligand complexes that will be generated, we have used one of the acceptable strategies by re-docking the inhibitor on the 3D crystal structures for PTP1B (PDB ID: 1T49) and α-glucosidase (3TOP). The ability of the docking tools to regenerate binding poses of co-crystallized inhibitors found in the X-ray or NMR structures was tested [42]. As shown in Figure 2, superimposition of docked and co-crystallized ligands suggests that our docking tools conveniently regenerated both the binding configuration and the molecular interaction in a comparable manner to the crystal structures. Root mean square deviation (RMSD) values of 0.943 and 0.981 obtained for α-glucosidase and PTP1B co-crystallized ligands respectively (Fig. 2) proved that the docking method was reliable and acceptable to predict the binding mode of the compounds evaluated.
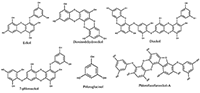
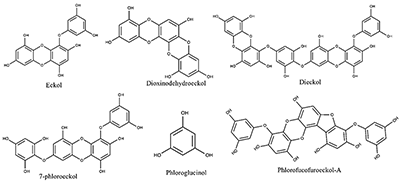
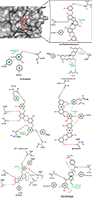
Exploring Interaction of the Seaweed-Derived Antidiabetic Compounds with PTP1B Six compounds used in this study for the ligand-binding experiment are shown in Figure 3. They are bioactive ingredients isolated from marine algae exhibiting antidiabetic activities; however, the mechanisms of the antidiabetic activities have not be exhaustively investigated. PTP1B remains one of the diabetes-related and most widely studied phosphatases with 121 structures so far deposited in the protein databank. Previous studies, using PTP1B knockout animals revealed that inhibitors of the PTP1B might address obesity and DM type 2 [4,6].
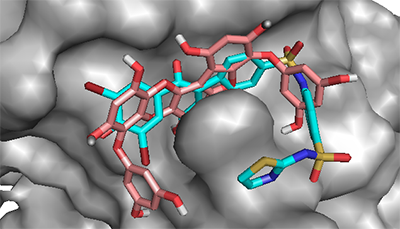
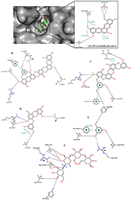
This explains why PTP1B has evolved as an attractive therapeutic target for the management of these human diseases. Thus, we have employed available in silico tools to investigate the binding modes and order of molecular interaction of these seaweed-derived constituents with the protein. The structural analysis of the human PTP1B has shown that the protein has more than one binding pocket where ligands can interact (Fig. 1). Potential inhibitors could bind either at the active site or the allosteric site. Therefore, we first attempt to determine the specific binding location of each of the seaweed bioactive compounds on the PTB1B crystal structure. Identifying the specific binding cavity suitable for a ligand on PTP1B can provide insight into the particular mechanism involved in the inhibition, either competitive inhibition type or allosteric type of inhibition. We have therefore adopted the blind docking approach where we chose a box size that completely covered the protein structure for the molecular docking experiment. As such, the ligand is permitted to search for its most favorable binding pocket on the protein. According to the results obtained, all the ligands favourably and more preferably docked to the allosteric site relative to the active site. Accordingly, the seaweed components studied here are predicted as potential antidiabetic drug candidates. As shown in Figure 4, the comparable binding configuration was observed between the control ligand and the marine alga-derived ligands.
For comparative reasons, we further docked all the compounds at the active site of the protein. Table 1 shows the estimated binding energy values for each ligand at both active site and allosteric site. The ligands interacted with the allosteric site with the lesser ∆G value in comparison to the active site. Binding energy usually denotes docking affinity of ligands to a particular receptor where a low binding energy value represents a better binding and vice versa [43]. Hence, our results indicate the higher affinity of seaweed constituents to the allosteric site of PTP1B compared to the substrate binding site. The binding energy at the allosteric site also showed good correlation with experimental results (IC50) for the ligands (Table 1).
The detailed analyses of interaction between the protein and the antidiabetic ligands are shown in Table 2. The docking experiments revealed that the top ranked conformations of eckol and dieckol were well accommodated inside the putative allosteric site. These compounds are phlorotannins commonly found in the brown marine algae [23,44]. They contribute, among other constituents, to the therapeutic activities of the seaweeds [44]. Herein, dieckol established three hydrogen bonds with residues Arg199, Phe196, and Glu276 while eckol formed five hydrogen bonds with Ala189, Glu276, Gly277, Glu200, and Asn193. A common π-π interaction was observed with Phe196 and Phe280 for both compounds (Table 2).
Hydrophobic interactions also contribute to the stability of the macromolecular complex between PTP1B and the seaweed compounds. The higher affinity estimated for dieckol in the current study may be attributed to its extended structure compared to eckol, which permits the compound to conveniently occupy the putative binding pocket. Interestingly, dieckol has previously been reported for its antagonistic activity on PTP1B with the IC50 value of 1.18 µM compared to eckol with IC50 of 2.64 µM [23]. This shows the agreement between our in silico results and wet experimental results. Phloroglucinol has shown the least number of hydrogen bonds and highest binding energy value in the current study. Compared to dieckol, phloroglucinol interacted with PTP1B less efficiently and inhibited the enzyme with very low potency (IC50 = 55.48 µM) [23]. However, this reduced potency is in agreement with the lower affinity (-4.8 kcal/mol) obtained for the compound. This observation is expected since phloroglucinol is the precursor of the phlorotannins. As polyphenolic compounds, the compounds are actually synthesized via polymerization of their aromatic precursor phloroglucinol (1,3,5-trihydroxybenzene) and are characterized by a wide range of molecular weights and sizes. In addition, they have been identified as promising bioactive ingredients in brown seaweeds with possible health benefits in slowing down the progression of type 2 diabetes as well as the management of its associated complications [45]. In this study, we report high binding affinity of phlorofucofuroeckol-A to the allosteric pocket of PTP1B (Table 1). This result suggests the potent antidiabetic activity of the seaweed bioactive ingredient via inhibitory potential on PTP1B. Serendipitously, this observation is compatible with earlier reports obtained in in vitro experiments [23,46]. As clearly depicted on proteinsplus server and presented in Figure 5, all the compounds chosen for evaluation in the current study displayed requisite potential to interfere with the PTP1B enzymatic function. The molecular interactions are similar to that of the control ligand. The seaweeds-derived antidiabetic compounds shared hydrogen bonds, hydrophobic and π-stacking interactions with common amino acid residues at the allosteric binding pocket (Fig. 5). These residues are essential for PTP1B inhibition [7]. Excitedly, the potency of some of the bioactive compounds from seaweed such as dieckol was higher than that of the control ligand [23]. The molecular interaction as well as the binding energy values lend credence to their higher potency. Since the binding poses are comparable to that of known allosteric inhibitor («inhibitor 3») [7], the seaweed bioactive compounds are allosteric inhibitors of the PTP1B. In addition, it must be emphasized that the affinity of the compounds to the PTP1B allosteric pocket is relatively higher than that of the active site and, the interaction analysis has provided insight to the possible reasons for the differences in their IC50 (potency) against the protein.
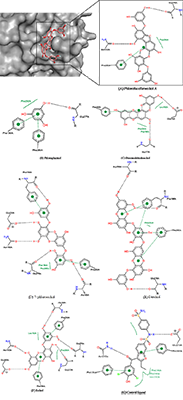
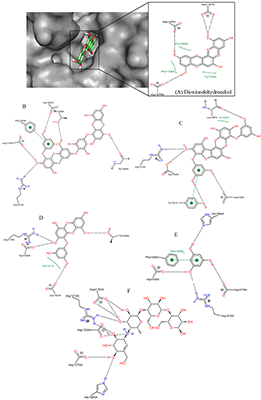
Interaction of α-Glucosidase (Maltase) with Seaweed-Derived Antidiabetic Compounds Figure 6 shows the binding pattern of dioxinodehydroeckol, a potent antidiabetic component of marine alga, compared to that of acarbose, a known α-glucosidase inhibitor [46]. Both compounds penetrated deep into the active site of the enzyme with potential to hinder substrate binding and its catalytic conversion. The aromatic rings of acarbose and dioxinodehydroeckol relatively are aligned, showing the seaweed-derived compound sitting very deep within the putative binding pocket and forming hydrogen bonds with Asp1157, Asp1420, and Asp1279 (Table 3).
This result suggests that seaweeds inhibit α-glucosidase mostly through the competitive inhibition mechanism. Compared to acarbose with binding energy value -8.7 kcal/mol, dioxinodehydroeckol possesses lower affinity (-8.3 kcal/mol). The higher potential of acarbose to block α-glucosidase catalytic function compared to the alga bioactive compound is consistent with their IC50 values of 9.9 µM [47] and 34.6 µM [23], respectively. Acarbose has been exhaustively studied and is currently being used in the control of postprandial hyperglycemia. Although the compound does not form hydrophobic interaction with interact α-glucosidase at the active site (Fig. 7), it is known to establish extensive hydrogen bonds, which are believed to contribute to its tight binding to the enzyme.
Among all the marine alga-derived compounds investigated in this study, phlorofucofuroeckol-A possessed the highest affinity to α-glucosidase with binding energy -10.5 kcal/mol, this result strongly agrees with its potent antidiabetic effects observed in wet experiments [23,48]. Accordingly, phlorofucofuroeckol-A is more potent than acarbose [49] with the energy value -8.7 kcal/mol. The polyphenolic compound is composed of numerous free hydroxyl (OH) groups and seven aromatic rings which play key roles in its interaction with the protein target. The presence of hydrophobic interaction with Trp1369 as well as a π-π interaction with the aromatic moieties of Tyr1251 in addition to hydrogen bonds with Asp1157, Arg1510, Tyr1251, Asp1420, and Leu1367 (Fig. 7) may be one of the reasons for the enhanced potency against α-glucosidase compared to acarbose. It is believed that such unique molecular binding patterns displayed by the algae-derived bioactive compounds on the active site of α-glucosidase might explain the subtle differences in their potency. Similar to acarbose, phloroglucinol lacks hydrophobic interactions with the protein. However, the compound formed π-stacking interaction with Phe1559 and hydrogen bonds with four amino acid residues including His1584, Asp1526, Arg1510, and Asp1279. As expected, the binding energy of phlorolgucinol is -5.4 kcal/mol, and affinity is higher than that of PTP1B. These results are compatible with results of in vitro studies on phlorolgucinol inhibition of α-glucosidase with IC50 141.18 µM [23]. 7-Phloroeckol is another antidiabetic compound from brown marine algae [23,35,48]. 7-Phloroeckol potently blocked α-glucosidase activity in vitro (IC50 = 6.13 µM) [23]. In the current study, we found the compound having high affinity (-9.2 kcal/mol) to the active site of the enzyme and forming hydrophilic interactions with Asp1526, Asp1420, Arg1510 and Gly1365. Despite the lack of π-π interaction, the 7-phloroeckol-glucosidase complex is characterized by hydrophobic interaction involving Met1421 (Table 3). Appreciable inhibitory potency was observed for eckol (-8.7 kcal/mol) and dieckol (-9.6 kcal/mol) corresponding to their IC50 value of 11.16 µM and 10.8 µM respectively [23]. Eckols are characterized by the presence of two dibenzodioxin units linked by a phenoxyl substitution at C4 while the dieckols comprise of two eckols as monomers, linked in a «head-to-tail» manner. The latter dispayed extensive hydrogen bond interactions and a single π-stacking interaction with α-glucosidase residues at the active site. Dieckol has been reported as one of the effective antidiabetic compounds found in seaweeds [25,49,50]. The predicted affinity and molecular interaction observed for eckol and dieckol in this study are consistent with their inhibitory potency. Occupying and barricading the active site, the bioactive compounds from seaweed can prevent substrate access to α-glucosidase for catalytic conversion [51]. The molecular weight, number of aromatic rings and number of OH groups are crucial to the varied interaction mechanism and inhibitory potential of the phlorotannins against their protein targets. Data of this study notably corroborate previous studies and validate seaweeds as great source of antidiabetic candidates [24,45,52] and thus, can be recommended for application in the management/prevention of DM and its associated complications. CONCLUSION In this study, α-glucosidase and PTP1B were investigates as targets for bioactive components of seaweeds as diabetes-related proteins to unravel their mechanism of interaction using computational procedures. The polyphenolic compounds from marine algae (phlorotannins) bind tightly to the allosteric pocket of PTP1B compared to the active site. Hence, the ligands studied in the current work are noncompetitive with respect to substrate binding on PTP1B. On the other hand, the bioactive compounds displayed competitive inhibition type against α-glucosidase by docking well into the putative substrate-binding site with tendency to interfere with substrate access, binding and conversion. Just like acarbose, the marine alga-derived constituents can block the degradation of complex carbohydrates into glucose and prevent the potential elevation of postprandial blood glucose level in DM type 2. All the compounds investigated in this study showed appreciable potential to inhibit the enzymes. The molecular size and number of OH groups on the polyphenolic compounds are essential for their inhibitory activities on PTP1B and α-glucosidase. They are hence proved suitable in the management of DM and its complications. Among the compounds however, phlorofucofuroeckol-A is the most potent compound with antidiabetic effect through high affinity and inhibition of both α-glucosidase and PTP1B. Overall, the interaction analysis performed in this study is crucial in providing plausible explanations for the variances in the potency (IC50) of the seaweeds against the enzymes. ACKNOWLEDGEMENTS We wish to acknowledge the technical support and encouragement received from all researchers at the Centre for Bio-computing and Drug Development (CBDD), Adekunle Ajasin University, Akungba-Akoko, Ondo state, Nigeria. REFERENCES
|