Prediction of Progestin Affinity for the Human Progesterone Receptor Based on Corrected RBA Data
Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 119121 Russia; *e-mail: a.v.mikurova@ibmc.msk.ru
Keywords: progesterone receptor; affinity; molecular docking; molecular dynamics; MM-PBSA
DOI: 10.18097/BMCRM00080
The modeling of complexes of 3 sets of steroid and nonsteroidal progestins with the ligand-binding domain of the nuclear progesterone receptor was performed. Molecular docking procedure, long-term simulation of molecular dynamics and subsequent analysis by MM-PBSA (MM-GBSA) were used to model the complexes. Using the characteristics obtained by the MM-PBSA method two data sets of steroid compounds obtained in different scientific groups a prediction equation for the value of relative binding activity (RBA) was constructed. The RBA value was adjusted so that in all samples the actual activity was compared with the progesterone activity. The third data set of nonsteroidal compounds was used as a test. The resulted equation showed that the prediction results could be applied to both steroid molecules and nonsteroidal progestins.
|
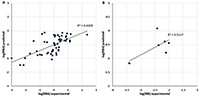
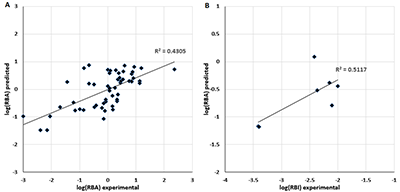
Figure 3.
The comparison of predicted RBA values with experimental RBA values in learning and RBI values in prediction (B).
|
|
CLOSE

|
Table 1.
The structure and experimental affinity value of the compounds used towards the progesterone receptor (see figs. 1-2). Selection of a particular variant of the ligand-receptor complex after docking.
|
|
CLOSE

|
Table 2.
The parameters of the linear regression equations obtained during learning and test results.
|
ACKNOWLEDGEMENTS
This work was performed within the framework of the Program for Basic Research of State Academies of Sciences for 2013-2020. The software tuning on hybrid cluster was supported by the Russian Foundation for Basic Research (project 18-29-03100).
REFERENCES
- Lagarde, N., Delahaye, S., Jérémie, A., Ben Nasr, N., Guillemain, H., Empereurmot, C., ... & Zagury, J. F. (2017). Discriminating agonist from antagonist ligands of the nuclear receptors using different chemoinformatics approaches. Molecular Informatics, 36(10), 1700020. DOI
- de Ziegler, D., & Fanchin, R. (2000). Progesterone and progestins: applications in gynecology. Steroids, 65(10-11), 671-679. DOI
- Lundgren, S. (1992). Progestins in Breast Cancer Treatment: A Reveiw. Acta Oncologica, 31(7), 709-722.. DOI
- Bursi, R., & Groen, M. B. (2000). Application of (quantitative) structure–activity relationships to progestagens: from serendipity to structure-based design. European journal of medicinal chemistry, 35(9), 787-796.Hillisch A., von Langen J., Menzenbach B., Droescher P., Kaufmann G., Schneider B., Elger W. (2003) Steroids, 68, 869-878. DOI
- Hillisch, A., von Langen, J., Menzenbach, B., Droescher, P., Kaufmann, G., Schneider, B., & Elger, W. (2003). The significance of the 20-carbonyl group of progesterone in steroid receptor binding: a molecular dynamics and structure-based ligand design study. Steroids, 68(10-13), 869-878. DOI
- Fedyushkina I.V., Skvortsov V.S., Romero Reyes I.V., Levina I.S. (2013). Molecular docking and 3D-QSAR on 16a,17a-cycloalkanoprogesterone analogues as progesterone receptor ligands, Biomeditsinskaya Khimiya, 2013, vol: 59(6), 622-635. DOI
- Cramer, R. D., Patterson, D. E., & Bunce, J. D. (1988). Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. Journal of the American Chemical Society, 110(18), 5959-5967. DOI
- Massova, I., & Kollman, P. A. (2000). Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspectives in drug discovery and design, 18(1), 113-135.Van Helden, S. P., Hamersma, H., & Van Geerestein, V. J. (1996). In Genetic Algorithms in Molecular Modeling, 159-192. DOI
- Van Helden, S. P., Hamersma, H., & Van Geerestein, V. J. (1996). Prediction of the progesterone receptor binding of steroids using a combination of genetic algorithms and neural networks. In Genetic Algorithms in Molecular Modeling (pp. 159-192). DOI
- Söderholm, A. A., Lehtovuori, P. T., & Nyrönen, T. H. (2006). Docking and three-dimensional quantitative structure− activity relationship (3D QSAR) analyses of nonsteroidal progesterone receptor ligands. Journal of Medicinal Chemistry, 49(14), 4261-4268. DOI
- SYBYL-X 2.1. Certara, Princeton, NJ, USA.
- Halgren, T. A. (1996). Merck molecular force field. II. MMFF94 van der Waals and electrostatic parameters for intermolecular interactions. Journal of Computational Chemistry, 17(5–6), 520-552. DOI
- Berman, H., Henrick, K., & Nakamura, H. (2003). Announcing the worldwide protein data bank. Nature Structural and Molecular Biology, 10(12), 980.
- Kuntz, I. D., Blaney, J. M., Oatley, S. J., Langridge, R., & Ferrin, T. E. (1982). A geometric approach to macromolecule-ligand interactions. Journal of Molecular Biology, 161(2), 269-288. DOI
- Case, D. A., Cheatham, T. E., Darden, T., Gohlke, H., Luo, R., Merz, K. M., ... & Woods, R. J. (2005). The Amber biomolecular simulation programs. Journal of Computational Chemistry, 26(16), 1668-1688. DOI
- Mikurova, A. V., Skvortsov, V. S., & Raevsky, O. A. (2018). Computational Evaluation of Selectivity of Inhibition of Muscarinic Receptors M1-M4. Biomedical Chemistry: Research and Methods, 1(3), e00072. DOI
- Federal Research Center Computer Science and Control of Russian Academy of Sciences [Electronic resource]: site. - Moscow: FRC CS RAS.- URL: http://hhpcc.frccsc.ru (application date: 09/12/2018)