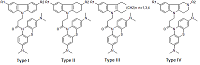
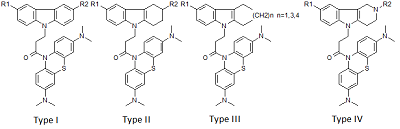
Investigation of NMDA Receptor Channel Blockers in a Series of Methylene Blue Conjugates Using QSAR and Molecular Modeling
1Institute of Physiologically Active Compounds of the Russian Academy of Sciences,
1 Severnyi pr., Chernogolovka, 142432 Russia; *e-mail: beng@ipac.ac.ru
2Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 119121 Russi
Keywords:NMDAR; channel blockers; QSAR; docking
DOI:10.18097/BMCRM00091
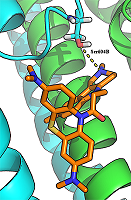
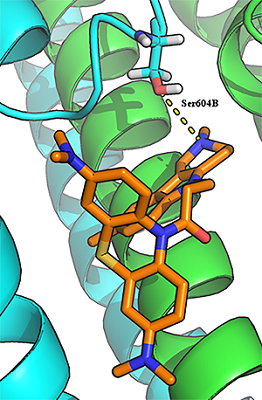
29 conjugates of methylene blue and four chemical structures, including derivatives of carbazole, tetrahydrocarbazole, substituted indoles and γ-carboline, combined with a 1-oxopropylene spacer have been studied as channel blockers of the NMDA receptor (binding site of MK-801) by using four QSAR methods (multiple linear regression, random forest, support vector machine, Gaussian process) and molecular docking. QSAR models have satisfactory characteristics. The analysis of regression models at the statistical level revealed an important role of the hydrogen bond in the complex formation. This was also confirmed by the study of modeled by docking complexes. It was found that the increase in the inhibitory activity of the part of compounds could be attributed to appearance of additional H bonds between the ligands and the receptor.
|
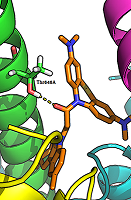
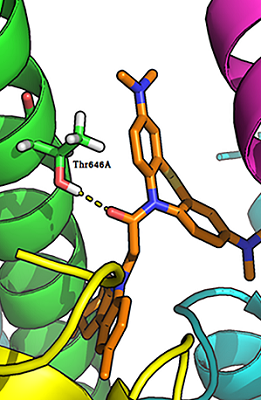
Figure 2.
The hydrogen bond between the carbonyl group of a ligand (compound 17) and the side chain of Thr646А-GluN17.
|
|
CLOSE

|
Table 1.
The formulas, biological activity (IC50mkM) and descriptors (α (Å3), Σ(Ca), Σ(Ca)/α, Eamax) of investigated compounds.
|
|
CLOSE

|
Table 1.
Statistical characteristics of QSAR models of NMDA receptor blockade.
|
|
CLOSE

|
Table 3.
The numbers of compounds, their experimental (Aexp), calculated (Acalc) activity values, and differences (Δ= Aexp - Acalc) of them in test set.
|
ACKNOWLEDGEMENTS
The work was performed within the framework of the state task of the IPAC RAS for 2019 (topic number 0090-2019-0004).
REFERENCES
- Wimo, A., Jonsson, L., Bond, J., Prince, M., Winblad, B. (2013). The worldwide economic impact of dementia 2010. Alzheimer`s Dement., 9(1), 1-11.e3. DOI
- Wimo, A., Guerchet, M., Ali, G-C., Wu, Y-T., Prina, A.M., Winblad, B., Jonsson, L., Liu, Z., Prince, M. (2017). The worldwide costs of dementia 2015 and comparisons with 2010. Alzheimer`s Dement., 13(1), 1-7. DOI
- Raevsky, O.A., Mukhametov, A., Grigorev, V.Y., Ustyugov, A., Tsay, S-C., Hwu, R.J-R., Yarla, N.S., Barreto, G.E., Aliev, G., Bachurin, S.O. (2018). Applications of Multi-Target Computer-Aided Methodologies in Molecular Design of CNS Drugs. Curr. Med. Chem., 25(39), 5293-5314. DOI
- Querfurth, H.W., LaFerla, F.M. (2010). Alzheimer's disease. New Engl. J. Med., 362, 329-344. DOI
- Carreiras, M. C., Mendes, E., Perry, M. J., Francisco, A. P., Marco-Contelles, J. (2013). The multifactorial nature of Alzheimer's disease for developing potential therapeutics. Curr. Top Med. Chem., 13, 1745-1770. DOI
- Liu H., Wang L., Lv M., Pei R., Li P., Pei Z., Wang Y., Su W., Xie X.-Q. (2014). AlzPlatform: An Alzheimer’s Disease Domain-Specific Chemogenomics Knowledgebase for Polypharmacology and Target Identification Research. J. Chem. Inf. Model., 54, 1050-1060. DOI
- Zambre, V.P., Hambarde, V.A., Petkar, N.N., Patel, C.N., Sawant, S.D. (2015). Structural investigations by in silico modeling for designing NR2B subunit selective NMDA receptor antagonists. RSC Adv., 5, 23922-23940. DOI
- Chtita, S., Larif, M., Ghamali, M., Bouachrine, M., Lakhlifi, T. (2015). Quantitative structure–activity relationship studies of dibenzo[a,d]cycloalkenimine derivatives for non-competitive antagonists of N-methyl-d-aspartate based on density functional theory with electronic and topological descriptors. J. Taibah Univ. Sci., 9(2), 143-154. DOI
- Elmbarki, C., Elhallaoui, M. (2017). Quantum calculations to construct a 3D-QSAR model based on PCP-TCP derivatives and Molecular Docking with NMDA receptor. J. Mater. Environ. Sci., 8(4), 1391-1400.
- Peters, O.M., Connor-Robson, N., Sokolov, V.B., Aksinenko, A.Yu., Kukharsky, M.S., Bachurin, S.O., Ninkina, N., Buchman, V.L. (2013). Chronic Administration of Dimebon Ameliorates Pathology in TauP301S Transgenic Mice. J. Alzheimer’s Dis., 33(4), 1041-1049. DOI
- Zhao, M., Liang, F., Xu, H., Yan, W., Zhang, J. (2016). Methylene blue exerts a neuroprotective effect against traumatic brain injury by promoting autophagy and inhibiting microglial activation. Mol. Med. Rep., 13, 13-20. DOI
- Raevsky, O.A., Trepalin, S.V., Grigorev, V.Yu., Yarkov, A.V., Bachurin, S.O. (2017) Certificate of state registration of the database "Multitarget organic compounds with potential effects on the central nervous system" № 2017620020.
- Coughenour, L.L., Barr, B.M. (2001). Use of Trifluoroperazine Isolates a [3H]Ifenprodil Binding Site in Rat Brain Membranes with the Pharmacology of the Voltage-Independent Ifenprodil Site on N-Methyl-D-aspartate Receptors Containing NR2B Subunits. J. Pharmacol. Exp. Ther. 296, 150-159.
- Raevsky, O.A., Grigor'ev, V.Yu., Kireev, D.B., Zefirov, N.S. (1992). Complete Thermodynamic Description of H-Bonding in the Framework of Multiplicative Approach. Quant. Struct.-Act. Relat. 11, 49-63. DOI
- Forsythe, G.E., Malcolm, M.A., Moler, C.B.(1977). Computer Methods for Mathematical Computations. Englewood Cliffs, NJ: Prentice-Hall, 270.
- URL: http://www.stat.berkeley.edu/~breiman/RandomForests/cc_examples/prog.f
- URL: https://github.com/jbcolme/fortran-ls-svm
- Obrezanova, O., Csányi, G., Gola, J.M.R., Segall, M.D. (2007). Gaussian Processes: A Method for Automatic QSAR Modeling of ADME Properties. J. Chem. Inf. Model., 47, 1847-1857. DOI
- Mitra, I., Saha, A., Roy, K. (2010). Exploring quantitative structure–activity relationship studies of antioxidant phenolic compounds obtained from traditional Chinese medicinal plants. Mol. Simul. 36(13), 1067-1079. DOI
- Trott, O., Olson, A.J. (2010). AutoDockVina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem., 31(2), 455-461. DOI
- Morris, G.M., Huey, R., Lindstrom, W., Sanner, M.F., Belew, R.K., Goodsell, D.S., Olson, A.J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem., 30(16), 2785-2791. DOI
- Kiralj, R., Ferreira, M.M.C. (2009). Basic Validation Procedures for Regression Models in QSAR and QSPR Studies: Theory and Application. J. Braz. Chem. Soc. 20(4), 770-787. DOI
- Gramatica, P. (2007). Principles of QSAR models validation: internal and external. QSAR. Comb. Sci. 26(5), 694-701. DOI
- Bolshakov, K.V., Gmiro, V.E., Tikhonov, D.B., Magazanik, L.G. (2003). Determinants of trapping block of N-methyl- D -aspartate receptor channels. J. Neurochem., 87, 56-65. DOI
- Beteringhe, A., Filip, P., Tarko, L. (2005). QSAR study for diarylguanidines, noncompetitive NMDA receptor antagonists. A new topological index AAd derived from local invariants of the chemical graphs of diarylguanidines. Arkivoc., 45-62. DOI
- Huettner, J.E., Bean, B.P. (1988). Block of N-methyl-D-aspartate-activated current by the anticonvulsant MK-801: Selective binding to open channels. Proc. Natl. Acad. Sci. USA., 85, 1307-1311. DOI