Generalized Predictive Model of Estimation of Inhibition of Muscarinic Receptors M1-M5
1Institute of Biomedical Chemistry, 10 Pogodinskaya Street, Moscow, 119121, Russia
2Institute of Physiologically Active Compounds, 1 Severniy pr., Chernogolovka, 142432, Russia; *e-mail: a.v.mikurova@ibmc.msk.ru
Keywords: acetylcholine muscarinic receptors; inhibitors; comparative inhibition; docking; computational methods; molecular dynamics; QSAR
Keywords: acetylcholine muscarinic receptors; inhibitors; comparative inhibition; docking; computational methods; molecular dynamics; QSAR
DOI:10.18097/BMCRM00129
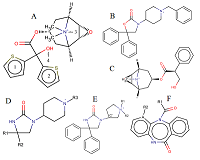
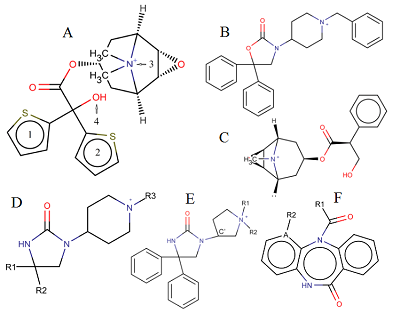
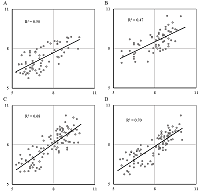
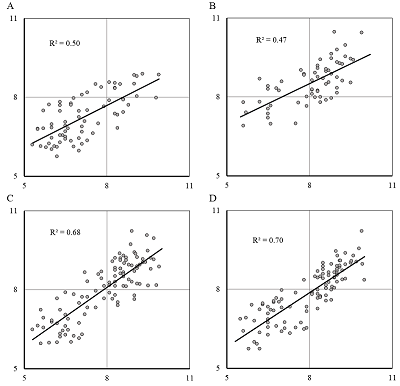
A general predictive model for assessing the inhibition constant (Ki) value of human acetylcholine muscarinic receptors M1-M5 by potential ligands has been constructed. We used information on the three-dimensional structure of human M1, M2, M4, and M5 receptors, as well as a model of the M3 receptor constructed according to homology based on the structure of the rat M3 receptor. A set of complexes of known inhibitors with the target receptor constructed by means of molecular docking, was selected using an additional option: the coincidence of the spatial position of 4 pharmacophore points of a tested inhibitor and tiotropium, for which the position in the crystal structure was known. For five types of M receptors 199 complexes with known Ki values were selected. Based on the data obtained during molecular dynamics simulation of these complexes by means of the MM-PBSA/MM-GBSA methods, their energy characteristics were calculated. They were used as independent variables in linear regression equations for pKi value prediction. The R2 prediction for the generalized equation was 0.7, and the mean prediction error was 0.55 logarithmic units with a range for pKi.= 4.7.
|
CLOSE

|
Table 2.
The parameters of training and testing linear regression models on sets for individual receptors and their combinations.
|
FUNDING
The work was carried out within the framework of the state task for 2018 (topic number 0090-2017-0020). The software porting on hybrid Power 9 cluster was supported by the Russian Foundation for Basic Research (project 18-29-03100).
SUPPLEMENTARY
Supplementary materials are available at http://dx.doi.org/10.18097/BMCRM00129
REFERENCES
- Eglen, R. M. (2006). Muscarinic receptor subtypes in neuronal and non-neuronal cholinergic function. Autonomic and Autacoid Pharmacology, 26(3), 219-233. DOI
- Langmead, C. J., Watson, J., & Reavill, C. (2008). Muscarinic acetylcholine receptors as CNS drug targets. Pharmacology & therapeutics, 117(2), 232-243. DOI
- Freedman, S. B., Dawson, G. R., Iversen, L. L., Baker, R., & Hargreaves, R. J. (1993). The design of novel muscarinic partial agonists that have functional selectivity in pharmacological preparations in vitro and reduced side-effect profile in vivo. Life sciences, 52(5-6), 489-495. DOI
- Mikurova, A., Skvortsov, V., & Raevsky, O. (2018). Computational Evaluation of Selectivity of Inhibition of Muscarinic Receptors M1-M4. Biomedical Chemistry: Research and Methods, 1(3), e00072. DOI:10.18097/BMCRM00072>/li>
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., ... & Bourne, P. E. (2000). The protein data bank. Nucleic acids research, 28(1), 235-242. DOI
- Vuckovic, Z., Gentry, P. R., Berizzi, A. E., Hirata, K., Varghese, S., Thompson, G., van der Westhuizen, E.T., Burger, W.A.C., Rahmani, R., Valant, C., Langmead, C.J., Lindsley, C.W., Baell, J.B., Tobin, A.B., Sexton, P.M., Christopoulos, A., Thal D.M. (2019). Crystal structure of the M5 muscarinic acetylcholine receptor. Proceedings of the National Academy of Sciences, 116(51), 26001-26007. DOI
- Thal, D. M., Sun, B., Feng, D., Nawaratne, V., Leach, K., Felder, C. C., ... & Kobilka, T. S. (2016). Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature, 531(7594), 335. DOI
- Haga, K., Kruse, A. C., Asada, H., Yurugi-Kobayashi, T., Shiroishi, M., Zhang, C., ... & Kobayashi, T. (2012). Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature, 482(7386), 547. DOI
- Kruse, A. C., Hu, J., Pan, A. C., Arlow, D. H., Rosenbaum, D. M., Rosemond, E., ... & Shaw, D. E. (2012). Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature, 482(7386), 552. DOI
- SYBYL-Х 2.1. Certara, Princeton, NJ, USA.
- Dei, S., Bellucci, C., Buccioni, M., Ferraroni, M., Guandalini, L., Manetti, D., ... & Romanelli, M. N. (2007). Synthesis, affinity profile, and functional activity of muscarinic antagonists with a 1-methyl-2-(2, 2-alkylaryl-1, 3-oxathiolan-5-yl) pyrrolidine structure. Journal of medicinal chemistry, 50(6), 1409-1413. DOI
- Peretto, I., Forlani, R., Fossati, C., Giardina, G. A., Giardini, A., Guala, M., ... & Raveglia, L. F. (2007). Discovery of diaryl imidazolidin-2-one derivatives, a novel class of muscarinic M3 selective antagonists (Part 1). Journal of medicinal chemistry, 50(7), 1571-1583. DOI
- Peretto, I., Fossati, C., Giardina, G. A., Giardini, A., Guala, M., La Porta, E., ... & Santoro, E. (2007). Discovery of diaryl imidazolidin-2-one derivatives, a novel class of muscarinic M3 selective antagonists (Part 2). Journal of medicinal chemistry, 50(7), 1693-1697. DOI
- Scapecchi, S., Marucci, G., Matucci, R., Angeli, P., Bellucci, C., Buccioni, M., ... & Teodori, E. (2001). Structure–activity relationships in 2, 2-diphenyl-2-ethylthioacetic acid esters: unexpected agonistic activity in a series of muscarinic antagonists. Bioorganic & medicinal chemistry, 9(5), 1165-1174. DOI
- Kuntz, I. D., Blaney, J. M., Oatley, S. J., Langridge, R., & Ferrin, T. E. (1982). A geometric approach to macromolecule-ligand interactions. Journal of molecular biology, 161(2), 269-288. DOI
- Case, D. A., Cheatham, T. E., Darden, T., Gohlke, H., Luo, R., Merz, K. M., ... & Woods, R. J. (2005). The Amber biomolecular simulation programs. Journal of computational chemistry, 26(16), 1668-1688. DOI
- Massova, I., & Kollman, P. A. (2000). Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspectives in drug discovery and design, 18(1), 113-135. DOI
- Federal Research Center Computer Science and Control of Russian Academy of Sciences [Electronic resource]: site. - Moscow: FRC CS RAS.- URL: http://hhpcc.frccsc.ru (application date: 03/09/2020)