Construction of a Chimeric Human Gene Encoding Renalase with a Modified N-Terminus
Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 10119121 Russia; * e-mail: valfed38@yandex.ru.
Keywords:PCR, cloning, expression, renalase, signal sequence
DOI:10.18097/BMCRM00137
Renalase (RNLS) is a recently discovered protein that plays different roles inside and outside cells. Extracellular RNLS exhibits protective effects on the cell, acting on its receptor proteins, while intracellular RNLS acts as FAD-dependent oxidoreductase (EC 1.6.3.5). The ratio of the intracellular and extracellular forms of this protein, as well as the mechanisms and factors responsible for its transport from the cell, remain unknown. One of the approaches to studying these issues can be the creation of chimeric forms of this protein with modified fragments of its amino acid sequences. This work describes a method for constructing a chimeric human RNLS gene encoding RNLS without its N-terminal peptide.
INTRODUCTION
In order to evaluated a role of the N-terminal peptide of renalase we have developed a chimeric human RNLS gene encoding RNLS without its N-terminal peptide and expressed it in both procaryotic and eukaryotic systems.
MATERIALS AND METHODS
Construction of the chimeric human RNLS gene encoding RNLS without its N-terminal peptide was based on the exon linkage of coding sequences described earlier [13, 14].
RESULTS
Table 1 shows primers used for PCR
|
CLOSE

|
Table 1.
Primers used in PCR
|
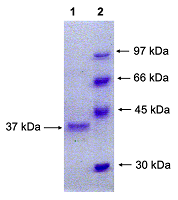
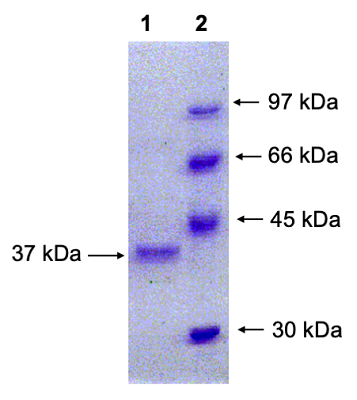
Sequence analysis of all studied clones resulted in identification of nucleotide substitutions (see Fig. 1); they were located in the third position of the following codons: gcC-Ala, atC-Ile, and tcG-Ser (Table 2). Due to codon, the amino acid sequence of the chimeric RNLS protein remained unchanged. These mutations detected in the RNLS gene could be attributed to the previously constructed pET-hRenI vector [14].
|
CLOSE

|
Table 2.
Degeneracy of codons for alanine (Ala), isoleucine (Ile), and serine (Ser)
|
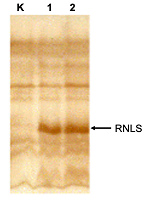
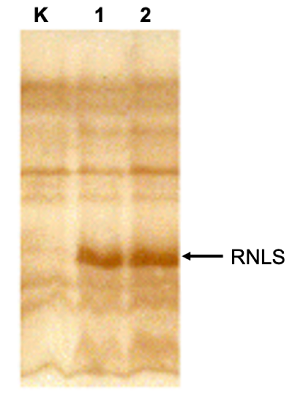
Mass spectrometric analysis of cell extracts, performed using a synthetic human RNLS proteotypic peptide labeled with stable isotopes of 13C and 15N [20], also confirmed the presence of the chimeric protein in transfected cells (and its absence in control, nontransfected cells).
Cloning of the RNLS gene and expression of the recombinant protein was supported by the Russian Foundation for Basic Research (project no. 20-015-00104). Mass spectrometric studies were performed within the framework of the Basic Research Program of the State Academies of Sciences for 2013-2020.
This work does not contain any results of research conducted with the participation of humans or the use of animals as objects.
The authors declare no conflicts of interest.
REFERENCES
- Xu J., Li G., Wang P., Velazquez H., Yao X., Li Y, Wu Y., Peixoto A., Crowley S., Desir G.V. (2005) Renalase is a novel, soluble monoamine oxidase that regulates cardiac function and blood pressure. J. Clin. Invest, 115(5), 1275–1280. DOI
- Medvedev A.E., Veselovsky A.V., Fedchenko V.I. (2010) Renalase, a new secretory enzyme responsible for selective degradation of catecholamines: achievements and unsolved problems. Biochemistry (Moscow), 75(8), 951-958. DOI
- Baroni S., Milani M., Pandini V., Pavesi G., Horner D., Aliverti A. (2013) Is renalase a novel player in catecholaminergic signaling? The mystery of the catalytic activity of an intriguing new flavoenzyme. Curr. Pharm. Des., 19, 2540-2551. DOI
- Desir G.V., Peixoto A.J. (2014) Renalase in hypertension and kidney disease. Nephrol. Dial. Transplant., 29(1), 22-28. DOI
- Moran G.R. (2016) The catalytic function of renalase: A decade of phantoms. Biochim. Biophys Acta, 1864(1),177-186. DOI
- 6.Wang Y., Safirstein R., Velazquez H., Guo X.J., Hollander L., Chang J., Chen T.M., Mu J.J., Desir G.V. (2017) Extracellular renalase protects cells and organs by outside-in signalling. J. Cell Mol. Med., 21(7), 1260-1265. DOI
- Kolodecik T.R., Reed A.M., Date K., Shugrue C.A., Patel V., Chung S.L., Desir G.V., Gorelick F.S. (2017) The serum protein renalase reduces injury in experimental pancreatitis. J. Biol. Chem. 292(51), 21047–21059. DOI
- Wang L., Velazquez H., Chang J., Safirstein R., Desir G.V. (2015) Identification of a receptor for extracellular renalase. PLoS One, 10, e0122932. DOI
- Moran G.R., Hoag M.R. (2017) The enzyme: Renalase. Arch. Biochem. Biophys., 632, 66-76. DOI
- Milani M., Ciriello F., Baroni S., Pandini V., Canevari G.,Bolognesi M., Aliverti A. (2011) FAD-binding site and NADP reactivity in human renalase: a new enzyme involved in blood pressure regulation. J. Mol. Biol., 411(2), 463-473. DOI
- Fedchenko V.I., Buneeva O.A., Kopylov A.T., Veselovsky A.V., Zgoda V.G., Medvedev A.E. (2015) Human urinary renalase lacks the N-terminal signal peptide crucialfor accommodation of its FAD cofactor. International Journal of Biological Macromolecules, 78, 347–353. DOI
- Fedchenko V., Kopylov A., Kozlova N., Buneeva O., Kaloshin A., Zgoda V., Medvedev A. (2016) Renalase Secreted by Human Kidney НЕК293Т Cells Lacks its N-Terminal Peptide: Implications for Putative Mechanisms of Renalase Action. Kidney Blood Press Res., 41, 593-603. DOI
- Fedchenko V.I., Kaloshin A.A. (2019) A simplified method for obtaining cDNA of low-copy and silent eukaryotic genes using human renalase as an example. Biomedical Chemistry: Research and Methods. 2(2), e00101. DOI
- Fedchenko V. I., Kaloshin A.A., Mezhevikina L.M., Buneeva O.A., Medvedev A.E. (2013) Construction of the Coding Sequence of the Transcription Variant 2 of the Human Renalase Gene and Its Expression in the Prokaryotic System. Int. J. Mol. Sci. 14, (6), 12764-12779. DOI
- Lee, P.Y., Costumbrado, J., Hsu, C.Y., Kim, Y. H. (2012) Agarose Gel Electrophoresis for the Separation of DNA Fragments. J. Vis. Exp., 62, e3923, DOI
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature; 227, 680-685. DOI
- Kushner, S. R. (1978). An improved method for transformation of Escherichia coli with ColE1-derived plasmids. In: Genetic engineering (Boyer H.B. and Nicosia S., eds.), p. 17, Elsevier/North-Holland, Amsterdam.
- Cohen, S.N., Chang, A.C.Y., Hsu, L. (1972). Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Esherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA, 69, 2110-2114.
- Gallagher S., Winston S.E., Fuller S.A., Hurrell J. G.R. (2011) Immunoblotting and Immunodetection. Current Protocols in Cell Biology 52 (1), 6.2.1-6.2.28.
- Kopylov A.T., Fedchenko V.I., Buneeva O.A., Pyatakova N.V., Zgoda V.G., Medvedev A.E. (2018) A new method for quantitative determination of renalase based on mass-spectrometry determination of a proteotypic peptide labeled with stable isotopes. Rapid Communications in Mass Spectrometry. 32(15), 1263-1270, DOI