|
CONTENTS |
Proteomics of the Human First Trimester Chorionic Villi Associated with Anembryonic Pregnancy
1Institute of Biomedical Chemistry; 10 Pogodinskaya St., Moscow, 119121 Russia;*e-mail: cyp450@mail.ru Key words: chorionic villi sampling; anembryonic pregnancy; tandem mass spectrometry; bioinformatics analysis; human chromosome 18 DOI: 10.18097/BMCRM00076 INTRODUCTION
Chorionic villus sampling (CVS) is the only diagnostic test available in the first trimester, which allows for diagnostic analyses between 10 and 14 weeks of gestation [1]. CVS is successfully used for a genome-wide non-invasive prenatal test of chromosome abnormalities including screens for trisomy 21, 18, 13 and sex chromosome aneuploidies [2]. Uncultured chorionic villi are the preferred samples for prenatal diagnosis of lysosomal storage disorders either by enzyme assays or by molecular studies [3,4]. In the post-genomics era, transcriptomics and proteomics approaches provide exhaustive information on protein expression as well as protein post-translation modifications and function under given conditions, to describe the complexity and functionality of the molecular mechanisms involved in the course of disease [5]. Although proteomics studies become now routine, there are not many works devoted to the analysis of chorionic proteins [6–8]. Two dimensional gel electrophoresis (2DE) followed by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF) was applied to reveal the proteomic profile of the CV cells [6, 9–10]. 2DE proteomics allowed to identify several proteins involved in the pathophysiological mechanisms. However, MALDI-TOF and 2DE is the most applicable for identification of highly abundant, large-molecular-weight proteins. At the same time shotgun MS fragmentation using collisional-induced dissociations (CID) or electron-transfer dissociation (ETD) provide direct amino acid sequences [8]. Some studies reported a gel-free tandem mass tags labeling-based proteomic analysis, which was used to identify differentially expressed proteins between CVS from normal pregnant women and early pregnancy loss [7,11]. Anembryonic pregnancy (blighted ovum, “empty sac”) is the most common identifiable pathology in the first trimester of pregnancy. It accounts for ∼15% of all first-trimester abortion [12]. Excluding those with chromosome abnormality, there remains a high percentage of abortion occurring without discernible mechanisms [13]. In this study, we have characterized the proteomes of CVS by using shotgun proteomic – high performance liquid chromatography connected with tandem mass spectrometry (LC-MS/MS) – to identify differentially abundant proteins between the two groups (blighted ovum and normal pregnancy). We performed bioinformatics analysis to determine the biological processes involved in the pregnancy pathology. MATERIALS AND METHODS Clinical samples Eight chorionic villus sampling from patients with the blighted ovum (the study group) and twenty four villus sampling from women with normal pregnancy (the control group) who underwent terminations of pregnancy due to psychological reasons at the same gestational age (6–11 weeks) were collected from the Center of Family Planning and Reproduction (Moscow, Russia). The diagnosis of blighted ovum was based on the transvaginal ultrasound results. CVS were collected via transcervical approach; fetal-derived chorionic villi were obtained under binocular magnifying glasses. Patient recruitment and sample collection protocols were approved by the local ethics rules of the Pirogov Russian National Research Medical University (Moscow, Russia) and the Center of Family Planning and Reproduction. Informed consent was obtained from all patients. All the samples were stored at −80°C until the use. Tissues were washed in ice-cold and sterile 0.9 % NaCl, dissected using a microscope to remove endometrial tissues and fetal membranes, and finally stored at −80°C until further analysis. Sample Preparation Among all the CVS collected, six normal CVS and two CVS with “empty sacks” were randomly sampled for subsequent analysis of chorionic proteome. A total of 100 mg of each CVS was homogenized and solubilized in six volumes of lysis buffer containing 7 M urea, 2 M thiourea, 65 mM dithiothreitol, and 1% of protease inhibitor E64 and kept for 30 min at 4°C with shaking on Vortex every 10 min. After sonication at 4°C using the program for the ultrasonic BANDELIN Sonopuls HD 2070 (Germany) (2 cycles for 50 s with a 5 s interval), lysed cells were centrifuged (Hettich zentrifugen micro 12-24, Germany) at 15000 rpm at room temperature for 15 min × 2 times to remove debris. Total protein concentration of solubilized CVS was determined by the Bradford assay [14], using bovine serum albumin as a standard. In-solution tryptic digestion The pair of CVS (175 µg of protein) for each study (n = 2) or control (n = 6) groups were in-solution digested as described in accordance with a standard protocol [15]. LC-MS/MS analysis Separation and identification of the peptides were performed on a Ultimate 3000 nano-flow HPLC ("Dionex", USA) connected to Orbitrap Exactive ("Thermo Scientific", USA) mass spectrometer equipped with a Nanospray Flex NG ion source ("Thermo Scientific") as described earlier [16]. Three independent LC-MS/MS runs were performed for each sample. Data processing Totally, 36 LC-MS/MS runs were carried out for all groups in raw format and analyzed using Progenesis LC-MS software ("Nonlinear Dynamics Ltd.", UK) as described previously [16]. Protein identification was performed in Mascot software with decoy [17] against SwissProt (SP, 2012_11 version, .fasta format) for Homo sapiens. Trypsin was specified as the proteolytic enzyme and up to one missing cleavages was allowed. Pyridylethylation (C) was used as static modification. Oxidation of methionine was set as variable modification. Charge states of +2, +3, and +4 were taking into account for parent ions. Mass tolerance was set to ± 10 ppm for parent ion masses and ± 0.05 Da for fragment ion masses; false discovery rate (FDR) ≤ 1%. The peptides identified in Mascot software with significance index (SI) > 13 were considered as significant. Data processing was performed using the R [software www.r-project.org]. Overrepresentation analysis against Gene Ontology biological processes was performed and visualized using the cluster Profiler library [18]. Only proteins identified by three or more peptides were used in the analysis. The UniProt IDs of differentially expressed proteins were converted into Entrez Gene ID. Obtained p-values of enrichment significance were adjusted according to Benjamini-Hochberg procedure [www.jstor.org/stable/2346101]. RESULTS The groups of patients included in the present study showed insignificant differences in pregnancy duration (9.1 ± 1.7 weeks and 9.3 ± 2.1 weeks in the control group and anembryonic group, respectively), and the number of previous pregnancies (in turn 2.2 ± 1.4 vs 1.6 ± 1.9). Although there was insignificant difference between the average age of patients in blighted ovum vs normal pregnancy, 36.0 ± 4.1 years and 29.8 ± 5.5 years, respectively, it seems that anembryonic pregnancy was more common with increasing age of the mother. This observation closely match those of the literature data [19]. Examination of the CVS proteome was performed by Progenesis LC-MS software ("Nonlinear Dynamics Ltd."). Two peak lists for control and study groups respectively were generated and exported for a Mascot MS/MS search. Three LC-MS/MS technical replicates were performed for each type of CVS to expand the portion of the identifiable proteins and the number of high quality peptides per protein. For CVS proteome analysis, we selected only identifications, which were made by multiple peptides (≥ 3). Proteins matched by one unique peptide were also considered when they were identified in all replicates. The shotgun approach revealed 3696 peptides, which represented 399 proteins of the control CVS. Among identified proteins, approximately 37% were represented only by a single peptide. For anembryonic CVS, a total number of 1866 peptides and 212 proteins were determined and 50% of these were represented by only one peptide. Total of 258 proteins were common for both studied groups, 260 were detected only in control CVS group and 73 proteins were detected only in anembryonic villi. Thus, 591 proteins in total were identified in solubilized fraction of CVS. Just 37% of all protein identifications were made by ≥ 3 high quality peptides and others were made by one or two peptides. For analysis of differences between the control and anembryonic CVS, a joint mgf file containing the feature list for protein identifications of both control and anembryonic pregnancy group was generated by the Progenesis LC-MS software. The list of proteins differently presented in anembryonic chorionic villus samples compared to control is shown in Table S1. Proteins with fold change > 1.5 were regarded as being differentially altered in the anembryonic CVS versus control pregnancy. This resulted in detection of the 187 differentially expressed proteins between the control and the blighted ovum; these included 53 up-regulated and 134 down-regulated proteins, respectively. Genes located on chromosomes 7, 16, and 18 mainly encoded proteins with increased content while genes of chromosomes 1, 2, and 11 mainly encoded proteins with decreased abundance. Proteins, encoded by genes located on chromosome 17, showed both up- and down-regulation in anembryonic pregnancy compared to normal gestation. The 20 most important up- and down-regulated proteins observed in the anembryonic CVS are listed in Table 1.
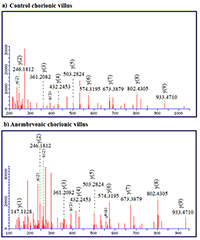
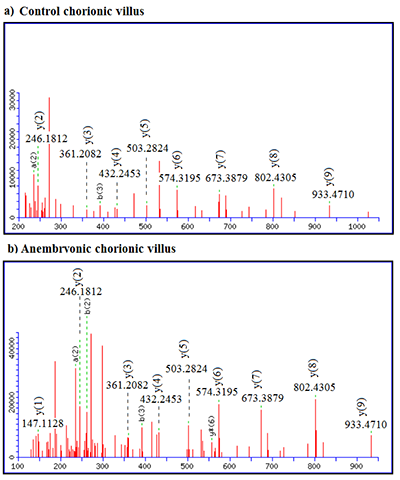
LC-MS/MS data showed that CVS from anembryonic pregnancy demonstrated the decline of the content of cytokeratin 7 (encoded by chromosome 12), which presented exclusively in trophoblast cells [20]. We also detected a decrease of ezrin (EZR) abundance (encoded by the gene of chromosome 6), its importance in human implantation was also demonstrated in [21]. Besides, our analysis revealed decreased abundances of creatine kinase B-type (CKB, encoded by the gene located on chromosome 14), required for tissues with high-energy demands. In addition we detected a decreased abundance of such proteins involved in glycolysis, as: glyceraldehyde-3-phosphate dehydrogenase (GAPDH, a key enzyme, encoded by chromosome 12), alpha-enolase (ENO1, encoded by chromosome 1), and phosphoglycerate kinase 1 (PGK1, encoded by the gene located on chromosome X). Some proteins implicated in the regulation of a large spectrum of both general and specialized signaling pathways exactly 14-3-3 protein zeta/delta (YWHAZ, encoded by the gene located on chromosome 8) also demonstrated decline of their normalized abundance in blighted ovum comparing to normal pregnancy. Proteins with ≥ 5-increased abundance were attributed to down-regulation of endothelial cell proliferation and drug metabolic process (ATP synthase subunit alpha, mitochondrial encoded by the gene located on chromosome 18) and response to epidermal growth factor (Major vault protein encoded by the gene located on chromosome 16). Among the proteins involved in the processes of apoptosis the proteins encoded by the genes located on chromosome 18 were identified (Table 1). These included down-regulated 3-ketoacyl-CoA thiolase, mitochondrial (ACAA2) and up-regulated cytosolic non-specific dipeptidase (CNDP2) and tyrosine-protein kinase Yes (YES1). Figures 1-2 serves as illustration of the up-regulated CNDP2 protein in anembryonic CVS (b) compared to control (a). We observed an increase in the sequence coverage of the protein: up to 59% in the blighted ovum compared to 33% in control. Furthermore the intensities of peptides spectra fragmentation (e.g. of peptide MMEVAAADVK (2+)), were also higher in the anembryonic CVS vs the control (Fig. 2).
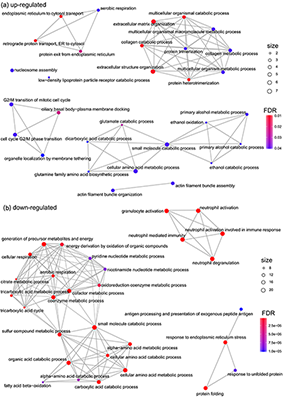
In order to characterize the proteome in anembryonic pregnancy in terms of biological processes we performed GO enrichment analysis of differentially expressed up- and down-regulated proteins by using the cluster Profiler library [18] (Fig. 3). According to the GO biological process, 53 up-regulated and 134 down-regulated proteins matched the database. We found that up-regulated proteins were overrepresented in the follows categories: small molecule catabolic processes,drug metabolic process and G2/M phase transition (Fig. 3a). The down-regulated proteins were significantly involved in tricarboxylic acid cycle, cellular respiration and amino acid metabolic processes (Fig. 3b).
The N-terminal acetylation is one of the most common protein modifications in eukaryotes [22]. A database search conducted with possible protein/peptide acetylation (Nt-acetylation) led to a decrease in the number of identified proteins in the control CVS as well in anembryonic samples (about 15% in both cases). Totally, 322 and 182 proteins were determined in control CVS and in “empty sac”, respectively. By implementing LC-MS/MS with both control and anembryonic samples, we identified 2122 peptides, which represented approximately 95 common proteins, and 15 peptides were acetylated (Table 2). Of the 11 acetylated peptides identified in total in anembryonic CVS, nine of them were common with acetylated peptides detected in control. Table 2 shows that there were four Nt-acetylated peptides – nos. 6, 12, 14 and 10 – absent (‘nd’) in the anembryonic chorionic villi but presented (‘+’) in the control. We also observed the reverse situation when the Nt-acetylated peptides were presented (‘+’) in the study group and absent (‘nd’) in the control. For example, the PTM-sensitive search did not reveal acetylated peptide of CNDP2.
DISCUSSION For screening birth defects or genetic disorders representing from 3% to 5% of all pregnancies many prenatal testing are used. These include serum and carrier screening, ultrasound, and amniocentesis; the goals of these tests are to identify women with pregnancies at high risk of chromosomal abnormalities or birth defects [1]. CVS remains the only diagnostic test available between 10 and 14 weeks of gestation; it is used for diagnostic analyses, including in situ hybridization fluorescence, karyotype, microarray, molecular testing, and gene sequencing. The proteomic profiling of CVS in combination with polymorphism analysis may enable us to find out particular molecular complexes or pathways in the birth defects or genetic disorders. Anembryonic pregnancy is the most common identifiable pathology in the first trimester of pregnancy [12]. Many reasons in relation to blighted ovum might be suspected; they may be associated with chromosomal abnormality, abnormal cell division, as well as poor quality of the sperm or the egg, acute viral and bacterial diseases in the early stages of gestation, endocrine disorders in a pregnant woman, and drug treatment with embryotoxic effect or chronic alcohol consumption. In this paper, we have analyzed the proteome alterations in parallel profiling of control and the anembryonic CVS by LC-MS/MS approach. We identified 53 up-regulated proteins and 134 down-regulated proteins in anembryonic CVS. Among the proteins with changed abundance three proteins encoded by chromosome 18 (CNDP2, YES1, and ACAA2, Table 1) are associated with apoptosis [23]. In addition, up-regulation of MVP was detected that could be important in the resistance of trophoblast cells to apoptosis [24]. Some proteins, such as GAPDH, ENO1, PGK1, and YWHAZ demonstrated the decrease in their content approximately by 7.5 times in the blighted ovum compared to the control group. Reduction in the content of these proteins was observed due to chronic beer consumption [25]. Decorin (DCN, encoded by the chromosome 12 gene), a leucine-rich proteoglycan produced by decidual cells, limits invasion and endovascular differentiation of extravillous trophoblast cells during early placentation. DCN with a more than 12-fold increase in normalized abundance in the anembryonic CVS was identified (Table 1); this could indicate poor endovascular differentiation of trophoblast cells in the blighted ovum. [26]. Therewith, we revealed down-regulation of EZR, which is an important for the maintenance of human cells size/mechanical properties and proliferation. We identified disequilibrium of acetylated proteins in anembryonic CVS, as compared to the control samples. The diverse functions of this modification have begun to be uncovered over the past decade; these include regulations of protein half-life, protein-protein and protein-membrane interactions, and subcellular localization as well the dysregulation of N-terminal modifying proteins at human disease including fetal problems [22,27,28]. The N-terminal acetylation of a specific protein can be either complete or partial, and in the latter case the protein exists in both acetylated and non-acetylated forms [29]. One example was the protein encoded by the gene located on chromosome 18, CNDP2; its modified form was recognized in the anembryonic CVS, while in the control samples the unmodified protein was detected (Table 2). As many proteins are N-terminally acetylated, it is expected that new functional roles of this modification in particular of proteins (especially those encoded by chromosome 18) will continue to emerge in the future. In conclusion, our results demonstrated the changes of the CVS proteomic profile in the pregnancy with the “empty sac”. We have detected the changes of various biological processes (Fig. 3a and 3b) that affect the trophoblast function and embryo development. In our opinion CVS can be a useful tool for understanding the role of proteins potentially and functionally involved in the different pathophysiological mechanisms underlying various biological status. Herein we have demonstrated involvement of proteins encoded by genes located on chromosome 18 in the pregnancy pathology, which will allow the chromosome-centric approach for better understanding of the normal and disease biology. ACKNOWLEDGEMENTS This work was performed within the framework of Fundamental Scientific Research Program of the Russian Academy of Sciences for 2013–2020. SUPPLEMENTARY Supplementary materials are available at http://dx.doi.org/10.18097/BMCRM00076 REFERENCES
|