|
Direct Detection of miRNAs miR-34a, -145, and -218 with CRISPR/Cas13a-Nuclease
Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 119121 Russia; e-mail: ryzhakova.olga@list.ru
Key words: microRNA; detection; CRISPR/Cas-nuclease DOI: 10.18097/BMCRM00203 INTRODUCTION Small non-coding RNA molecules known as microRNAs (miRNAs) play an important role in posttranscriptional regulation of gene expression [1, 2]. A miRNA is transcribed as a primary microRNA in the nucleus and exported to the cytoplasm as a precursor miRNA where it is processed into a mature miRNA of 18 to 25 nucleotide (nt) long [3]. Aberrations in miRNA expression were shown to closely associate with various diseases including cancer [2, 3]. Due to involvement of miRNAs in cellular regulatory processes, they are considered as promising biomarkers of pathological states, with a focus on detection of cell-free miRNAs circulating in body fluids [4, 5]. The common approach to detection of circulating miRNAs relies on polymerase chain reaction (PCR), using a combination of stem-loop and conventional primers [4, 6]. While PCR is performed in specialized laboratories and requires expensive equipment, presently there is a worldwide call for easy-to-use technologies, which can bring diagnostics closer to a final user – the format known as “point-of-care testing” (POCT) [7]. The emerging CRISPR-based molecular diagnostics relying on the use of CRISPR/Cas-nucleases per se or in combination with isothermal amplification methods promises the rapid, sensitive, and inexpensive detection of nucleic acids and appears to well suit the POCT requirements [8]. Among CRISPR/Cas-nucleases, Cas13a nuclease (further referred to as Cas13a for simplicity) forms a complex with the RNA molecule of a specific sequence – the so-called “guide RNA” (gRNA). The sequence of gRNA is composed of a part responsible for interaction with Cas13a (‘direct repeat’) and a part (28 nt long”spacer”) complementary to a target sequence (“protospacer”) [9]. Cas13a activates only if an almost perfect heteroduplex between spacer and protospacer is formed, resulting in a nicking of a target sequence. Upon activation, Cas13a additionally acquires the so called “collateral” or trans-activity, which is an ability of the nuclease to unselectively cleave, alongside with the target RNA, all RNA molecules present around [10]. Recently, Shan et al. [11] demonstrated that a miRNA could be directly detected by using complexes of Cas13a and gRNA with a spacer truncated down to 20 nt. In this work, we tested whether this approach can be applied to directly quantify three particular miRNAs, viz. miR-34a, miR-145, and miR-218. These miRNAs are shown to be involved in tumorigenesis of cervical cancer [12] and their molecular signature was suggested as a potential diagnostic and prognostic biomarker for this type of cancer [5]. MATERIALS AND METHODS Reagents All chemicals were purchased from “Merck” (USA), if not stated otherwise, and were of the ACS grade or higher. Solutions used were prepared with Milli-Q quality water (18 MΩ·cm). DNA oligonucleotides listed in Table 1 were chemically synthesized and purified by “Evrogen” (Russia). The recombinant Cas13a nuclease was expressed, purified, and characterized as described in details elsewhere [13, 14]. Synthesis of miRNAs and guide RNAs To synthesize miRNAs and gRNAs, DNA templates (Table 1) were equimolarly mixed with DNA oligonucleotide T7F (Table 1). Its sequence is complimentary to that of the promoter of T7 RNA polymerase (T7 promotor) and annealed by incubating the mixture at 95°C for 2 min, followed by cooling down to room temperature. The gRNA was synthesized using a TranscriptAid T7 High Yield Transcription Kit (“Thermo Fisher Scientific”, USA) according to the manufacturer instructions and purified by phenol/chloroform extraction followed by ethanol precipitation. The RNA pellets were air-dried and dissolved in nuclease-free water. RNA concentrations were measured on a NanoDrop 1000 spectrophotometer (“Thermo Fisher Scientific”). The lengths of synthesized RNAs met those expected as revealed by denaturing gel electrophoresis (data not shown). The aliquots of miRNAs and gRNAs were stored at -80°C until use.
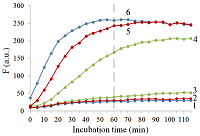
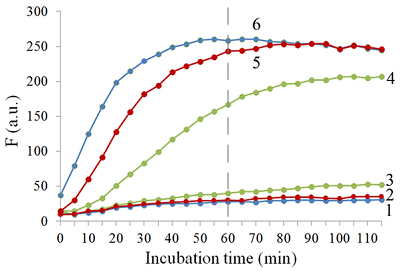
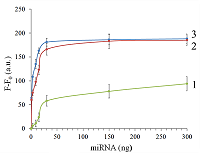
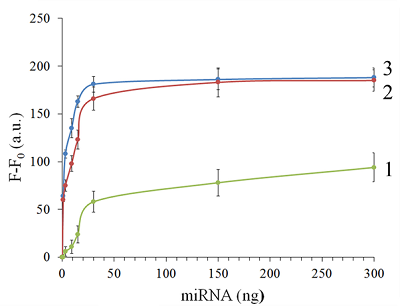
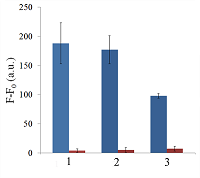
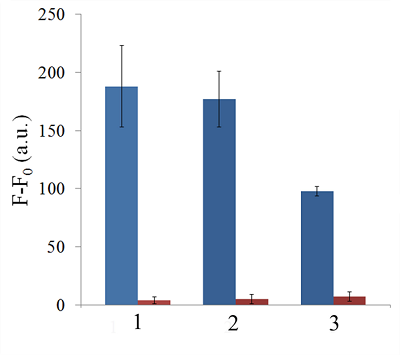
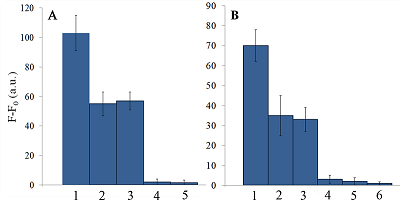
Isolation of total and small RNA from cultured cells The total RNA was isolated from XL1 Blue strain of E. coli with ExtractRNA reagent (“Evrogen”, Russia) and from HepG2 cultured cells with RNeasy Mini Kit (“Qiagen”, Germany) according to the manufacturers’ protocols. Small RNA (RNA of length < 200 nt) was prepared from the total RNA extracted from HepG2 cells with RNA Clean & Concentrator-25 Kit (“Zymo Research”, USA) following the manufacturer’s instruction. RNA concentrations were determined spectrophotometrically on a NanoDrop 1000 spectrophotometer. Aliquots of total and small RNA preparations in nuclease-free water were stored at -80°C until use. Measurement of collateral activity of Cas13a ribonuclease To measure the Cas13a collateral activity, RNaseAlert® Lab Test Kit v2 (“Thermo Fisher Scientific”) was used. The kit includes reporters which are short RNA molecules labelled with a fluorescent dye (presumably fluorescein) and a quencher (further referred to as FQ-reporters). Upon cleavage of a FQ-reporter, the fluorophore and the quencher separate; this results in an increase of fluorescence. The sample preparation was carried out as described earlier [14], with gRNA/Cas13a complexes formed at a molar ratio of 4:1. The TECAN Infinite 200 Pro plate reader (“TECAN”, Austria) was used to monitor fluorescence at 490 nm and 520 nm wavelengths for excitation and emission, respectively. Each 50-µL sample contained 650 ng of Cas13a and varying amounts of miRNAs tested in the absence or presence of either total or small RNA as a background. RESULTS AND DISCUSSION Figure 1 shows the kinetics of FQ-reporter cleavage by Cas13a in the presence and absence of target miRNAs. As seen, despite the truncation of a gRNA spacer length down to 21-23 nt, the gRNA/Cas13a complexes are indeed able to recognize targets and acquire the collateral activity. The values of fluorescence at 1 h incubation time where they reached or approached a plateau (Fig. 1) were chosen to further evaluate the detection of a target miRNA by a gRNA/Cas13a complex. Figure 2 demonstrates that Cas13a in the complex with the truncated gRNA does selectively recognize the corresponding target miRNA in the presence of two other miRNAs tested. The dependences of FQ-reporter cleavage on the amount of target present in the reaction mixture for each target miRNAs are shown in Figure 3. For two miRNAs, miR-218 and -34a, the dependencies were quite similar, with a fast increase and a further saturation at approximately 30 ng (corresponding to 4 pmol) of a target miRNA in the test sample. For miR-145, the fluorescence increase is also saturated at 30 ng of the target but at a substantially lower level (Figure 3). The low limit of detection (LOD) for miR-218 and miR-34a was estimated as ~1 ng per reaction (140 fmol per reaction = 80 nM in the reaction mixture), based on a signal-to-noise ratio of 3, while for miR-145 it was about 10 ng (1.4 pmol). The LODs for miR-218 and miR-34a are much lower than LOD for miR-145 though the latter is the longest miRNA out of three tested (21 and 22 nt vs. 23 nt, Table 1). Also, stability of the target/spacer heteroduplex (which can be affected not only by the oligonucleotide length but also by its sequence) was the highest for miR-145 among miRNAs tested, as estimated by calculating values of free energy with the RNAcofold tool available at http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAcofold.cgi (ΔG = -44.63, -34.64, and -42.08 kcal/mol for miR-145, -218, and -34a, respectively). Thus, the observed differences cannot be accounted for by simply assuming that the recognition of miR-145 by the corresponding truncated gRNA is less effective due to pure physical reasons. Furthermore, the best LOD value we obtained (140 fmol) was dramatically higher than that of 5 amol (that corresponded to 0.5-pM target concentration in the reaction mixture) reported by Shan et al. for the direct detection of miR-17 (23 nt long) [11]. To some extent, such dramatic differences can be explained by the fact that we performed our experiments under rather suboptimal conditions and a better detection sensitivity can be achieved via their optimization. Yet, it seems doubtful that the optimization could improve the sensitivity by more than 4 orders of magnitude. Besides that, under identical conditions, miR-34a, -218, and -145 demonstrated a 10-fold difference in the detection sensitivity. One cannot rule out that for miR-34a, -218, and -145 the sensitivity, even at optimal conditions, would remain much lower than that reported by Shan et al. for miR-17 [11]. It is thought that different spacer sequencies can somehow influence an acquiring of collateral activity by Cas13a, for example, by affecting the formation of a gRNA/Cas13a complex via unwanted partial pairing of the spacer with the gRNA repeat sequence and therefore decreasing the actual number of complexes. To evaluate how the Cas13a-based detection of a target miRNA would be affected by the presence of a large variety of RNA sequences, we have added 100 ng of total RNA either from bacterial (E. coli) or from mammalian (HepG2) cells to the reaction mixture containing 3 ng of miR-34a (Fig. 4A) or miR-218 (Fig. 4B). As seen, in both cases, the registered fluorescence signal decreased by approximately twice but still allowed to reliably detect the presence of target miRNA in the mixture. Interestingly, neither E. coli nor HepG2 total RNA per se produced appreciable fluorescence signals (Figure 4) thus confirming a high selectivity of gRNA/Cas13a complexes despite the gRNA truncation. However, after addition of 100 ng of small RNAs from HepG2 cells to the reaction mixture containing 3 ng of miRNA-218, no fluorescence signal was observed (Figure 4B). The diversity of RNA sequences is much higher in a preparation of small RNAs due to domination of ribosomal RNA (with a rather limited sequence variety) in total RNA. One may assume that short-living but multiple interactions of target RNAs and the spacers of corresponding gRNAs with sequences of the background RNA may be attributed to the observed decline in the detection effectiveness due to a partial base pairing. To some extent, this reminds the off-target effects for the case of small interfering RNAs (siRNAs) that is also a result of partial base pairing (e.g., [15, 16]). While an almost perfect base pairing of the gRNA spacer with a counterpart RNA sequence is necessary to activate Cas13a, the partial base pairing of gRNA spacer with surrounding RNA sequences can interfere with such pairing, leading to a decrease in an effective concentration of gRNA/Cas13a complexes. Moreover, the partial pairing of a target miRNA with the surrounding RNA sequences can also decrease its effective concentration by the same token. In overall, this can make the direct Cas13a-based detection of miRNAs in a complex medium the additionally sequence-dependent, providing acceptable sensitivity for ones [11] but not for other miRNAs. Alongside with direct detection of miRNAs with Cas13a, using the simple technique based on a cleavage of FQ-reporters to register the Cas13a collateral activity as in [11] and the present study, more sophisticated approaches were suggested where the additional signal amplification was employed. These approaches include the enhancement of the reporter cleavage signal by means of glucose oxidase mediated conversion of glucose with hydrogen peroxide formation [17, 18], by isothermal amplification methods such as catalytic hairpin assembly (CHA) [19] and exponential amplification reaction (EXPAR) [20], or by combining the reporter cleavage by Cas13a with subsequent activation of CRISPR/Cas-nuclease Cas12a or Cas14a [21, 22]. In the most cases, the outstanding values of LOD ranged from few zmol to amol per reaction were reported. Apparently, so high sensitivity can alleviate the problem of partial base pairing resulting in a decrease of the effective target concentration and potentially provide the detection of miRNA of any sequence in a complex medium. CONCLUSIONS In this study we have demonstrated the feasibility of direct detection of three miRNAs, miR-34a, -145, and -218. It is based on the registration of a cleavage of molecular reporters bearing a fluorophore and a quencher catalyzed by CRISPR/Cas13a-nuclease activated by the presence of a target miRNA. Among the miRNAs studied tested the detection sensitivity varied by 10-fold, presumably due to the unwanted intramolecular partial base paring of gRNA. The miRNA detection with Cas13a nuclease strongly depended on the presence of background RNA thus potentially compromising its direct application to complex media in a general case. Further optimization of measurement conditions including additional amplification of the signal generated by collateral activity of Cas13a nuclease is necessary for direct detection of miR-34a, -145, and -218 in biological samples. COMPLIANCE WITH ETHICAL STANDARDS This article does not contain any research involving humans or using animals as objects. FUNDING The study was performed within the framework of the Program for Basic Research in the Russian Federation for a long-term period (2021–2030) (No. 122030100170-5). CONFLICT OF INTEREST The authors declare no conflict of interest. REFERENCES
|